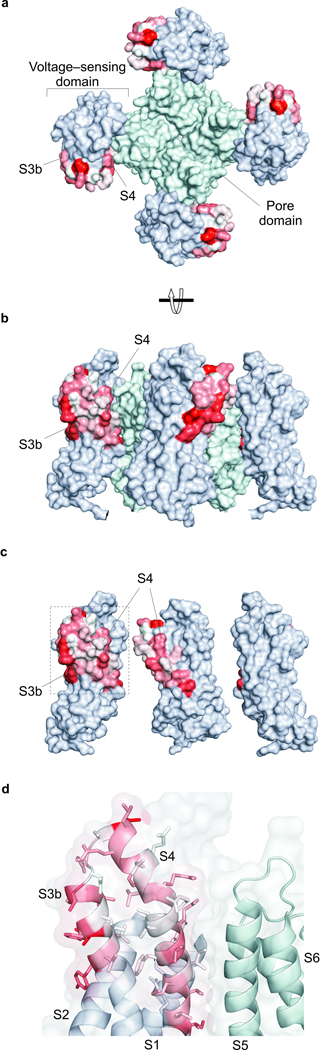
Figure 4.
Coupling energies mapped onto the X-ray structure of a Kv channel and a model for how lipid modification alters toxin affinity. (a) Surface representation of the Kv2.1/Kv1.2 paddle chimera with coupling energies mapped onto the paddle region, viewed from the extracellular side of the membrane. A color gradient between white and red was used to represent increasing |ΔG| values from 0 to 1.3 kcal mol−1. Residues in the pore domain are colored light cyan and those in the S1–S4 domains outside the paddle motif are colored light blue. (b) Surface representation of the Kv2.1/Kv1.2 paddle chimera viewed from the side. (c) Surface representation of the Kv2.1/Kv1.2 paddle chimera viewed from the side with front S1–S4 voltage-sensing domain and central pore domain removed. (d) Close up view of the S3b–S4 paddle motif with transparent surfaces and side chains colored as in a. Residues in the S1–S2 loop were removed for clarity. PDB accession code is 2R9R and all structures were drawn using PyMol (DeLano Scientific Inc.).