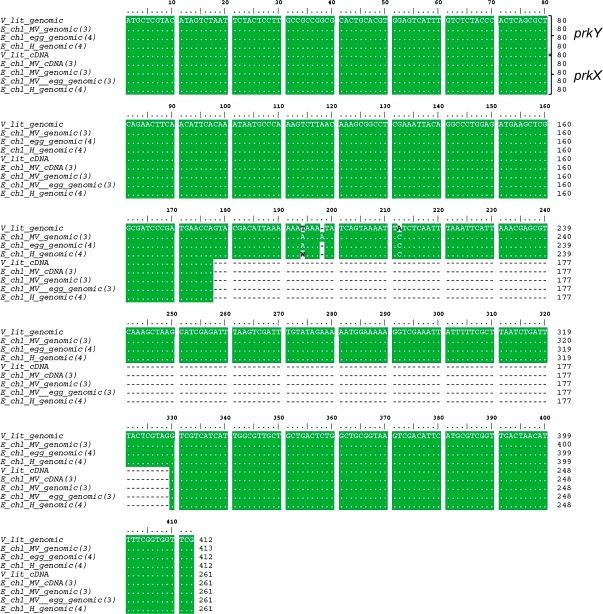
Figure 7.
Alignment of Partial prk Sequences from V. litorea and E. chlorotica.
Clustal W alignment of nucleotide sequences amplified from V. litorea cDNA and genomic DNA, E. chlorotica total DNA from Martha's Vineyard (MV) and Halifax (H) collections, E. chlorotica (MV) egg DNA, and E. chlorotica (MV) cDNA using PRK5F and PRK5R primers (see corresponding PCR bands in Figure 6A and 6B). Numbers in parentheses after the organism name refer to the number of sequences analyzed for each. Numbers at the end of each line indicate the respective nucleotide positions of the amplified product beginning with the start codon ATG. A dot indicates that the residues in that column are identical in all sequences in the alignment. A wavy dash (∼) marks the intron region. W = A or T. The larger, intron-containing sequence was labeled prkY, whereas the shorter sequence was labeled prkX.