Abstract
One of the major challenges in the genomic era is annotating structure/function to the vast quantities of sequence information now available. Indeed, most of the protein sequence database lacks comprehensive annotation, even when experimental evidence exists. Further, within structurally resolved and functionally annotated protein domains, additional functionalities contained in these domains are not apparent. To add further complication, small changes in the amino-acid sequence can lead to profound changes in both structure and function, underscoring the need for rapid and reliable methods to analyze these types of data. Phylogenetic profiles provide a quantitative method that can relate the structural and functional properties of proteins, as well as their evolutionary relationships. Using all of the structurally resolved Src-Homology-2 (SH2) domains, we demonstrate that knowledge-bases can be used to create single-amino acid phylogenetic profiles which reliably annotate lipid-binding. Indeed, these measures isolate the known phosphotyrosine and hydrophobic pockets as integral to lipid-binding function. In addition, we determined that the SH2 domain of Tec family kinases bind to lipids with varying affinity and specificity. Simulating mutations in Bruton’s tyrosine kinase (BTK) that cause X-Linked Agammaglobulinemia (XLA) predict that these mutations alter lipid-binding, which we confirm experimentally. In light of these results, we propose that XLA-causing mutations in the SH3-SH2 domain of BTK alter lipid-binding, which could play a causative role in the XLA-phenotype. Overall, our study suggests that the number of lipid-binding proteins is drastically underestimated and, with further development, phylogenetic profiles can provide a method for rapidly increasing the functional annotation of protein sequences.
Introduction
The three fundamental components of cells include proteins, lipids, and nucleotides. Proteins provide the machinery for lipid organization, storage, synthesis, and catalysis; thus, they have developed a vast array of functional domains capable of orchestrating these tasks. Further, lipids regulate a multitude of protein functions (protein-binding, enzymatic activity, trafficking, etc) and can even be integral to protein folding (e.g. proteins which contain lipids within their globular core). Therefore it is reasonable to consider that protein/lipid interactions are extremely common, and likely exist in most proteins. However, the number of proteins, either known or predicted to interact with lipids, is relatively small; only ~5% of proteins in the human proteome are annotated for the key word “lipid-binding” in the NCBI protein database.
The study of Zhu et al provides a clear demonstration that current computational methods for annotating lipid-binding function are insensitive (1). In this study, these authors identified 124 lipid-binding proteins from the yeast proteome using a high-throughput lipid-binding assay. Formal searches of these sequences using General Profile (GP) (2), Hidden Markov Model (HMM) (3–5), Support Vector Machine (SVM) (6), and Gene Ontology (GO) algorithms (7) predict lipid-binding for 3.23%, 3.23%, 12.9%, and 4.8% of this dataset, respectively. These results underscore the need for improved functional measures.
We recently proposed that phylogenetic profiles provide a unified framework for the study of structure, function, and evolution (8). A phylogenetic profile of a protein is a vector, where each entry quantifies the existence of the protein in a different genome (9–11), or the existence of an alignment within a profile knowledge-base (8). We and others have demonstrated that these approaches are applicable to whole molecule (Single Profile Method), to an isolated domain (Multiple Profile Method), and to single amino acids. Indeed, these methods have been used to infer protein function (8;10–12), protein structure (8), protein evolution (8;13), and even interaction partners (9). Despite these successes, phylogenetic profiles are a relatively untapped resource, and the accuracy and resolution which can be obtained with these measures has yet to be determined.
Towards this end, we present here a study which examines the ability of Single Amino Acid Phylogenetic Profiles (SAPPs) to identify the structural and functional determinants of lipid-binding within a structurally resolved set of SH2 domains. Our results demonstrate that, even at this nascent state, the Gestalt Domain Detection Algorithm Basic Local Alignment Tool (GDDA-BLAST) has the capacity to accurately predict lipid-binding domains (8;14–17). We demonstrate here that SAPPs provide more refined functional measurements for lipid binding. In support of this supposition, we examined a benchmark dataset of structurally resolved SH2 domains, some of which have been determined to bind lipids. Our results suggest that most, if not all, SH2 domains have lipid-binding capacity. Further, our analyses reveal that the SH2 domain of Tec family tyrosine kinases bind to lipid, and that simulation of XLA-causing mutations drastically alter the lipid-binding specificity and affinity of BTK. These simulations also isolate amino acids which are integral to, and surround the known phosphotyrosine and hydrophobic binding pockets. These data correlate well with the NMR study of Tokonzaba et al (18), which demonstrated that these pockets were involved in binding phosphatidylinositol (4,5) bisphosphate (PIP(4,5)2) by the SH2 domain contained in Abelson murine leukemia viral oncogene homolog 1 (c-abl), a distant relative of Tec kinases (18). Moreover, phylogenetic analysis of Tec kinase SH2 domains indicate that this region is evolving more rapidly than the other homologous domains contained in this family, suggesting this is a site of functional innovation. We envision that the methods presented here can be (i) extended to any functional class (e.g. nucleotide-binding, ATP-ase, phosphatase, etc), (ii) be harnessed to decode the most challenging protein datasets, and (iii) scaled up to screen proteomes and the vast quantities of sequences being obtained from metagenomic studies and other large scale sequencing projects.
Results
Generating Single Amino Acid Phylogenetic Profiles
Generating phylogenetic profiles using GDDA-BLAST begins by compiling a set of position specific scoring matrices (PSSM, i.e. domain profiles) that the query sequence is compared to (Fig 1a) (8). These profiles can be obtained from any protein-sequence knowledge-base source (e.g. Protein Data Bank (PDB), Pfam, SMART, NCBI Conserved Domain Database (CDD)) (2–4). In this study, we curated 131 profiles from CDD which are functionally related to peripheral lipid-binding (PLB) (8;17). These profiles contain multiple structural domains over a wide-range of lipid-binding specificity/affinity.
Figure 1. Single amino-acid phylogenetic profile generation and hierarchical clustering of structurally resolved SH2 domains.
(a)Workflow of using GDDA-BLAST to generate SAPPs using 131 profiles associated with peripheral lipid-binding activity. (i) For a query, embedded alignments are generated with each of 131 PLB profiles using GDDA-BLAST (i.e. reverse-COBBLER). These alignments are filtered using the thresholds of %identity and %coverage. (see Methods) A profile boundary in the query sequence is defined as overlapping the positive seeded alignments excluding seed over the query. (ii) By Smith-Waterman algorithm, the optimal local alignment is generated between a profile boundary region of query and a consensus sequence of PLB positive profile (i.e. a profile with at least one positive seeded alignment to the query). Based on the local alignment, each amino acid is scored as +2 for identities and +1 for positive substitution. For a query, the previous steps are repeated for every PLB profiles. Then, raw score at each amino acid is normalized by subtracting the average raw score of all amino acids. Finally, a SAPP (Single Amino-acid Phylogenetic Profile) of a query, which is a vector of normalized positional scores at each amino acid, is generated. (iii) The distribution of each amino acid and the number of amino acids with chemical properties (hydrophobic, positive charge and negative charge) from SAPP are incorporated into N (query) by M (compositional profile) matrix(31). Then, using the matrix, the query sequences are hierarchically clustered using Pearson’ correlation metrics.
(b) Dendrogram of SH2 domains hierarchically clustered using peripheral lipid-binding SAPPs. We observe 3 major clades in this dendrogram, all of which receive robust statistical support. We also observe that each clade contains SH2 domains which have been demonstrated to bind lipid experimentally. These results suggest that all of the SH2 domains tested contain lipid-binding activity.
Following this step, query sequences are embedded with a standard length of consensus sequence obtained from PLB PSSMs and then aligned to the parent PSSM using rps-BLAST(see (8;13) for a complete description). This embedding and alignment strategy is the reverse of the COBBLER algorithms developed by Henikoff and Henikoff (19;20). These authors demonstrated that embedding query sequences within PSSMs or consensus sequences rapidly and significantly improves the performance of multiple algorithms that employ PSSMs (e.g. PSI-BLAST, Smith-Waterman etc). GDDA-BLAST (i.e reverse-COBBLER) embeds PSSM consensus sequences within the query, thereby improving the performance of rps-BLAST (i.e. reverse PSI-BLAST).
From these results, each profile alignment above threshold is defined within the query sequence to create boundaries for our subsequent pairwise alignments. Next, to optimize our positional information, we re-align each profile above threshold with the query sequence using the Smith-Waterman algorithm (21). Raw scores for each residue are calculated by scoring a value=2 for identities and value=1 for positive substitutions from each alignment. The raw scores can be analyzed in multiple ways. For example, they can be plotted as a histogram to identify residues which are prominent in our measurements. Additionally, these results can be used to create an N (query) by M (compositional profile) matrix. The compositional profile is comprised of the 20 amino acids, plus three chemical classifications (hydrophobic, positive charge, and negative charge). These matrices can be used together to hierarchically cluster sequences that infer structural/functional relatedness. (see Methods for complete description).
SH2 Domains as a Model System for Lipid-binding
We chose to base our study on SH2 domains as (i) there are numerous structures for these domains in the PDB library, (ii) they are a prominent class of well-studied adaptor domains (22), and (iii) a select few have been determined to bind lipids experimentally (18;23); although, none are annotated for this function computationally (e.g. SMART, Pfam, InterProScan, CDD, SVM). The positive controls in this dataset include the lipid-binding SH2 domains from cabl, the p85 subunit of phosphoinositol-3-kinase (PI3K), and phospholipase C-γ2 (PLCγ2). We analyzed the 45 structurally resolved SH2 domains with GDDA-BLAST and plotted the results (see Methods and Supplemental Table 1). We observe many PLB peaks in regions of these proteins which are known to bind lipid (e.g. the catalytic core of PLCγ, the pleckstrin homology domain in SH2B adaptor protein 2 etc), the SH2 domains contained in these proteins, as well as other areas which have no annotation.
Following, we generated PLB SAPPs and created an N (SH2-domain) by M (compositional profile) matrix and performed hierarchical clustering with Pearson’s correlation. From these results, we observe three robust clades, all of which contain positive controls for lipid-binding. The correlation scores in this analysis are robust, and demonstrate that all of the SH2 domains in our analysis can be related using PLB SAPPs. Upon closer examination, we observe that three members of the tyrosine kinase expressed in hepatocellular carcinoma (Tec) Kinase family (BTK, bone marrow kinase gene on the X chromosome (BMX), and T and X cell expressed kinase (TXK)) cluster together, and are near both c-abl and PLC-γ2 in the dendrogram. Interestingly another family member, IL2-inducible T-cell kinase (ITK), lays in another clade, with its nearest neighbor spleen tyrosine kinase (Syk), a distant relative of ITK. These SH2 domains lie in close proximity to the N-terminal low-affinity lipid-binding SH2 domain of PI3K. Interestingly, in all cases of proteins containing two SH2 domains, the N-terminal SH2 domains are always separated from the C-terminal SH2 domains, suggesting that these domains are evolving distinct functionalities. Indeed, the C-terminal high-affinity lipid-binding SH2 domains of PI3K are clearly separated from the N-terminal SH2 domains, which might be expected based on the difference in their activity (23).
Modeling the Structural and Functional Determinants of the SH2 Domain of BTK
Based on the clustering of the Tec kinase family, we continued our computational analysis on these proteins. The Tec family of tyrosine kinases is composed of numerous members that are important to metazoan growth and development (24). Typically, these proteins are comprised of an N-terminal pleckstrin homology (PH) domain, followed by a Tec, SH3, SH2, and kinase domain (24). The PH domain in BTK is known to bind phosphatidylinositol (3,4,5)-trisphosphate during B-cell receptor stimulation. Further BTK’s SH2 domain can bind phosphorylated tyrosines such as the B-cell linker protein (BLNK). As both SH2 and SH3 domains have been demonstrated to also provide protein-protein interaction domains, it is speculated that these domains enable BTK to form multiple interactions within the B-cell receptor complex. Importantly, mutations in BTK that cause XLA have been identified in all of these domains (25).
Determining how these small mutations disrupt protein function is a daunting task. In general, unless a function has already been assigned to the protein domain containing the mutation, isolating a functional outcome is not feasible. Further, even if a function has been determined for the domain of interest, unknown secondary and tertiary functions can exist, making data analysis and interpretation quite difficult. For example, many of the XLA-mutations which have been tested experimentally do not alter the kinase activity of BTK significantly (25). In support of this observation, a study in a −/−BTK chicken B-cell line of 7 different XLA-mutations revealed only wild-type BTK could restore Ca2+ signaling in response to B-cell receptor stimulation (26). These data suggest that BTK contains multiple non-kinase activities that are required for proper function.
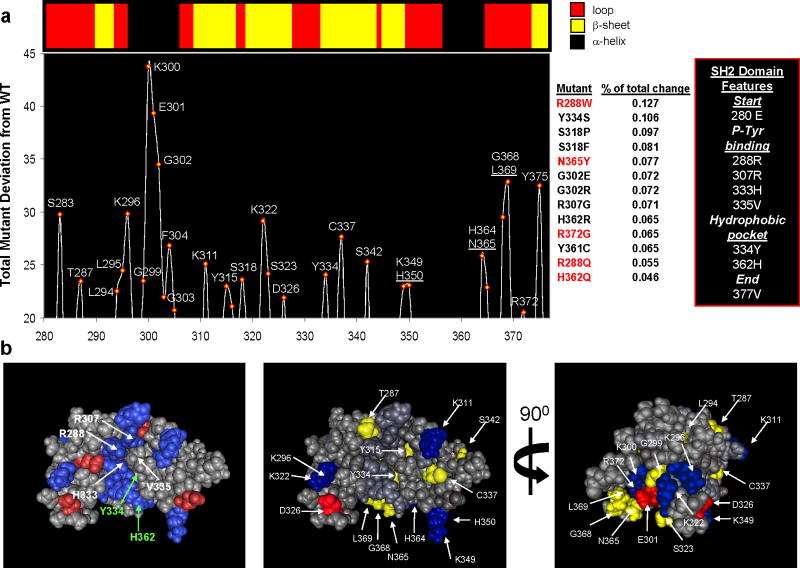
We previously demonstrated that GDDA-BLAST SAPPs can model the presence or absence of ATP-binding between closely related ankyrin repeats as well as amino acids that are chemically and structurally important to ATP binding (8). The same protocol also identifies lipid-binding in the catalytic pocket of serine racemase, and amino acids important for PIP2 binding (14). We wondered what effect XLA-mutations in the SH2 domain of BTK would have on our computational models. Initially, we simulated the 13 known XLA-causing mutations in the BTK SH2 domain and generated their SAPPs using the PLB. We were surprised to see that nearly all of the mutations tested (see Supplemental Table 1) altered our lipid-binding signals, with R288W (a mutation that inhibits phosphotyrosine binding), having the largest change in signal (Fig 2a right).
Figure 2. Phylogenetic profiles reveal putative lipid-binding residues in BTK.
(a) Top: Depiction of the secondary structural elements in the SH2 domain of human Bruton’s Tyrosine Kinase (PDB: 2GE9). Bottom: Positional analysis of human BTK for residues implicated in peripheral lipid-binding (PLB) from our phylogenetic profiles. The graph depicts the total change between sequences mutated to resemble 13 naturally occurring polymorphisms linked with XLA and wild-type sequence at each amino acid position (mutants in red were experimentally tested). Each sequence was compared with WT BTK and the absolute change recorded and finally summed for all mutations. The results indicate that the region proximal to the second phosphotyrosine-binding site (R307) is the site with the most change; however, positions throughout the p-Tyr and hydrophobic pocket can be identified. These predictions resemble those findings by Tokonzaba et al. in which they measured positions in the SH2 domain of c-Abl that display NMR structural perturbations in the presence of lipids (18)(see Supplemental Figure 1). (b) 3-D views of the SH2 domain of human Bruton’s Tyrosine Kinase (PDB: 2GE9). Left panel is colored for charged residues and labeled for those residues which make up the P-Tyrosine and Hydrophobic binding pockets. The middle and right panels are two views colored with those positions identified by GDDA-BLAST analysis. Residues colored in blue(−) and red(+) are charged.
Next, we compared the simulated SAPPs with the WT BTK sequence all-against-all to determine which amino acids signals changed the most in each simulation (absolute values, see Methods). The results plotted in Figure 2a reveal residues within the phosphotyrosine binding pocket and the hydrophobic pocket. These data are in excellent accord with the results of the study of Tokonzaba et al which demonstrate that both of these pockets are involved in phosphotyrosine-binding and lipid-binding by the SH2 domain of c-abl (Supplemental Figure 1a) (18). Further, many of the XLA-causing mutations themselves scored high in this analysis, in particular G302 and N365.
To ensure these results are not random, we performed Monte Carlo randomization of the SH3-SH2 domain sequence in BTK 105 times and repeated our GDDA-BLAST analyses (see Methods). We observe that, on average, the normalized PLB signal generated by this analysis has a constant rate of appearing at random (3.62 +/− 0.1 s.d.), which is well below what we observe using the WT-BTK sequence (Supplemental Table 1). We performed the same analysis using other regions of the protein (PH-domain, kinase domain) and obtained the same frequency. Thus, it appears that the alignments generated with the PLB have a relatively constant random frequency in any protein sequence. Next, we mapped the residues isolated in this analysis to the BTK SH2 domain structure (Fig 2b, Supplemental Figure 1b). The left panel displays a charge map of the domain, mapping the residues chemically involved in binding of tyrosine phosphorylated peptides from both pockets. The center and left panel display the residues isolated from our analysis. We observe that the strongest scoring residues from our analysis are close to the phosphotyrosine binding pocket in the amino-acid sequence, but spatially, most of these residues actually surround the hydrophobic pocket (left), not the phosphotyrosine binding pocket (right). These results suggest that the hydrophobic pocket is important to BTK SH2 domain lipid-binding.
SH2 Domains of Tec Kinases Bind Lipid
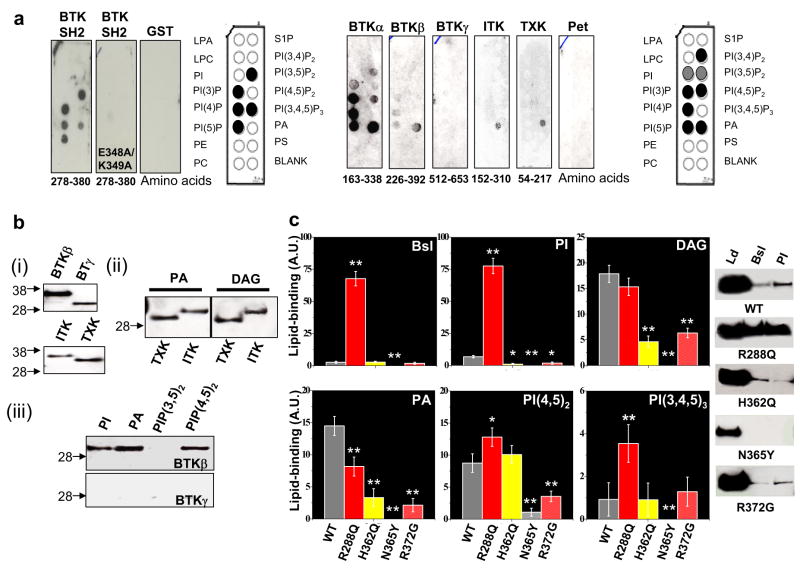
Armed with these results, we examined the lipid-binding capacity of Tec kinases with a series of in vitro functional assays. First, we performed PIP-strip© lipid binding assays (see Methods) using bacterially purified GST-tagged peptides of the SH2 domain from mouse BTK. We observe that WT-BKT-SH2 displays strong binding in these assays, while GST alone does not. We also prepared an E348A/K349A mutant, as these residues were prominent in our SAPP analysis (Fig 2a), and are charged residues that may participate in binding the negatively charged headgroup of phospholipids. We observe that these mutations completely abolish lipid-binding. Interestingly K349 lies in close proximity to H362 in the hydrophobic binding pocket (Fig 2b left). To assess whether the hydrophobic pocket was important to lipid-binding, we created a 6X-His-tagged peptide with the hydrophobic pocket deleted. To improve the folding of this construct, we included ~120 amino acids N-terminal to the BTK’s SH2 domains which contain an SH3 domain and the BTK motif (BTKα). In PIP-strip assays, this construct displays more robust and less-specific lipid-binding than the SH2 domain only (Fig 3a right). As the SH3 domain of BTK is also prominent in our PLB SAPPs (see Supplemental Table 1), we also made a construct which comprises the SH3 and SH2 domain (BTKβ). We also purified a region of the kinase domain in BTK, which is not predicted to bind lipids, to serve as a negative control (BTKγ). We observe that BTKβ binding is similar to BTKα binding, although not as robust while control preparations do not display lipid-binding. Peptides were also cloned from mouse ITK and mouse TXK for the region homologous to BTKβ. We observe that both ITK and TXK are specific for phosphatidic acid (PA) in this assay (Fig 3a right).
Figure 3. SH3-SH2 module of Tec Kinases bind lipids in vitro.
(a) Left: WT and mutant SH2 domains of mouse BTK were cloned and bacterially purified. These samples were tested for binding a PIP-strip(c) array by Western analysis with anti-GST. The results indicate that WT preparations bind inositol lipids with the mutant displaying significant reduction in binding. Right: Three additional fragments of BTK were tested: (1) BTKα: SH3 domain and SH2 P-Tyr binding pocket, (2) BTKβ: SH3 domain and whole SH2 domain, and (3) BTKγ: region inclusive to the small PLB signal observed in the kinase domain. Analogous peptides to BTKβ were made in mouse ITK and mouse TXK. All 5 peptides and negative control were assayed as above and probed with anti-His. The results indicate that BTKα, which lacks the hydrophobic pocket, displays different affinity/specificity when compared to BTKβ. Further, ITK and TXK are specific for phosphatidic acid (PA) in this assay. Neither BTKγ nor Pet control displayed strong binding. (b-i) Loading controls for bacterially purified protein. (b-ii) Liposomal assays of ITK and TXK demonstrate specific binding to PA and diacylglycerol containing vesicles (see Methods). (b-iii) Liposomal assays demonstrate that BTKβ but not BTKγ bind to liposomes containing PI, PA, or PIP(4,5)2. (c)b Quantitative liposomal binding assays for six different lipid compositions using WT BTKβ and naturally occurring XLA polymorphisms. On the right are representative Western analyses. The results indicate that these mutations in the P-Tyrosine binding pocket increase lipid binding affinity >50-fold in some cases while mutations in and near the Hydrophobic pocket have graded inhibition of lipid-binding.
To extend these findings, we next performed the more physiologically relevant liposomal lipid-binding assay (see Methods) (Fig 3b). Indeed, we observe that all peptides positive in the PIP-strip assays© also bound to liposomes. Interestingly, in addition to PA binding, BTK displayed some specificity for PIP(4,5)2 over PIP(3,5)2. Further, ITK and TXK were quite specific for PA and diacylglycerol (DAG) containing vesicles. These data demonstrate that the SH3-SH2 domain in Tec kinases is a lipid-binding module.
We then tested a number of lipid-mixtures to determine if lipid-binding capacity and/or specificity were altered (Fig 3c). When compared to WT control, all of the mutants tested (R288Q, H362Q, N365Y, R372G, and R288W [not shown]) had altered lipid-binding profiles. The R288Q (and R288W) displayed the largest increases in affinity and changes of specificity, in particular to liposomes containing phospholipids at similar ratios to the plasma-membrane and membranes containing PIP(3,4,5)3. Indeed, when the homologous mutation is made in c-abl, Tokonzaba and colleagues observed an increased affinity for PIP(4,5)2, consistent with this result (18). These mutants also have reduced affinity for PA and DAG, whose binding is conserved in all Tec kinases tested. Further, all of the mutations near the hydrophobic pocket severely inhibited PA and DAG binding, with the N365Y mutation being essentially devoid of all lipid-binding. Taken together, these results support the importance of the hydrophobic pocket in BTK lipid-binding as was suggested by our computational analysis (Fig 2). Moreover, it appears that the BTK has also evolved lipid-specificities which are distinct from its family members.
Phylogenetic Analysis of Tec Kinases
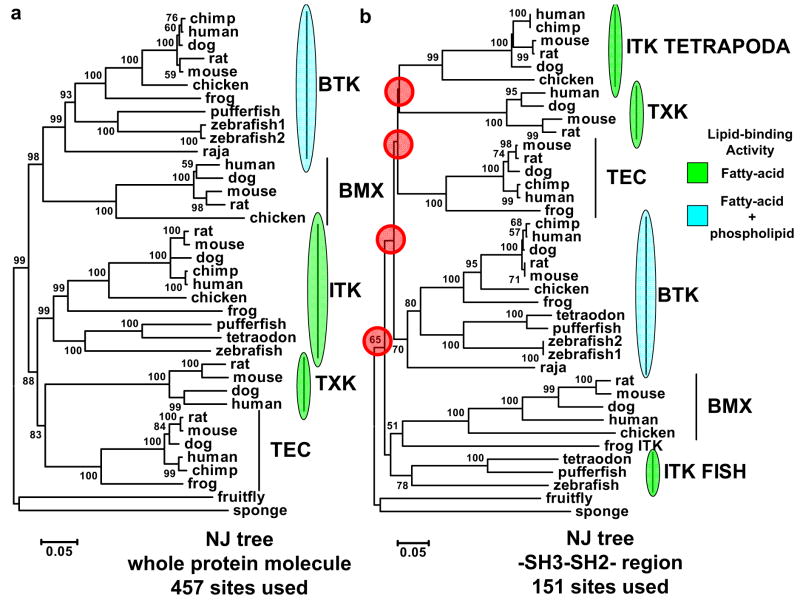
To place these results within an evolutionary context, we performed a phylogenetic study of the Tec kinases, using human and mouse sequences for each family member, with the Tec kinases from sponge and fruitfly serving as our outgroups (Fig 4a). We observe that Tec, ITK, TXK form the most ancient clade, followed by the BTK and BMX clade, all of which obtain robust statistical support. These clades also recapitulate the lipid-binding specificity we observe experimentally (Fig 3). To specifically investigate the SH3-SH2 domains of this family, we performed additional phylogenetic analysis using only this region of the family (151 sites). This tree’s topology is severely distorted and lacks statistical support at all deep branches (Fig 4b). Interestingly, the ITK sequences split into two clades and segregate to opposite ends of the tree in this analysis. This, coupled to the results from our hierarchical clustering (Fig 1b), suggest that the SH2 domain of ITK is distinct from the other Tec kinases, although this was not revealed by any functional assay in this study. The SH3-SH2 domain results are in stark contrast to trees generated using only the PH domain (92 sites) or kinase domain (227 sites) (Supplemental Figure 1c) which retain the same topology as the full length sequences (Fig 1a), as well as statistical support. Taken together, these results indicate that the SH3-SH2 domains in Tec kinases are diverging, and are likely an evolutionary site for functional innovation.
Figure 4. Phylogenetic Analysis of Tec Kinases.
(a) Phylogenetic analyses reveal SH3-SH2 domains in TEC Kinases are rapidly evolving. Neighbor-joining tree of full-length TEC, TXK, BMX, and BTK sequences from various taxa. This tree is rooted by the fruitfly and sponge TEC sequences. (b) Neighbor-joining tree using only the SH3-SH2 module of TEC Kinases as above. The results indicate that while full-length sequences (457 sites) provide a monophyletic tree with robust support, the tree generated from sites segregated to the SH3-SH2 module (151) lack proper topology and have little support. These results are not due to the reduced number of sites utilized since trees generated from PH domain only (92 sites) or SH1 domain only (227 sites) retain the full-length topology and have significant support (Supplemental Fig 1c).
Discussion
The results from this study demonstrate that single aminoacid phylogenetic profiles built from lipid-binding profiles can be used to accurately identify lipid-binding and residues of structural/functional importance within SH2 domains, which are not random. To our knowledge, no other computational algorithm can annotate lipid-binding function to SH2 domains or isolate key residues important for this function. This demonstrates the improved functional detection afforded by phylogenetic profiles. Further, the results from these analyses provided a rationale for our laboratory experimentation which led us to discover that: (i) Tec family kinase SH2 domains have lipid-binding capacity, which varies in affinity and specificity between family members, (ii) fatty-acid binding is common to all Tec family members, (iii) XLA-causing mutations in the SH2 domain of BTK all alter lipid-binding, and (iv) that the hydrophobic pocket of the BTK SH2 domain is critical for lipid-binding activity. These data imply that XLA-causing mutations in the SH3-SH2 domain of BTK may alter cellular functions related to lipid-binding such as trafficking, localization, protein-protein interactions, and/or activity. Thus, lipid-binding may play a causative role in the XLA-phenotype, which is important to consider when developing therapeutic strategies. It is also intriguing that functional measurements obtained by GDDA-BLAST can predict polymorphisms which alter protein functions, although the reliability of this method has yet to be rigorously evaluated.
An important observation from our study is the prediction that likely all SH2 domains bind lipid since the structurally resolved domains used in our study are from a variety of proteins. This additional functional annotation of SH2 domains allows for improved design of biological experiments which aim to determine their cellular functions (e.g. perhaps an observed functional defect is related to lipid-binding dysregulation not phosphotyrosine binding). Indeed, accurate annotation of lipid-binding in any protein would aid in designing appropriate experimentation. For example, most of the 124 proteins isolated from the Zhu study (1) are not predicted to bind lipid. When we analyzed these sequences using the random frequency of PLB signals in randomized BTK sequences as our baseline, our results predict regions in all of these proteins which bind lipid(s) (Supplemental Table 2). Thus, annotating proteomes for lipid-binding using this approach would provide important resource for the scientific community, and is one of our short-term goals. To that end we have submitted our molecular interaction data to IntAct, a public domain database [The protein interactions from this publication have been submitted to the IMEx (http://imex.sf.net) consortium through IntAct (pmid: 17145710) and assigned the identifier IM-11692. Direct link:http://www.ebi.ac.uk/intact/search/do/search?searchString=IM-11692].
In theory, the methods developed in this study can be applied to any protein functionality. In support of this theory, we have used this approach to identify multiple previously unidentified functions. For example, we identified ATP-binding in ankyrin repeats, as well as and lipid-binding/trafficking activity in the TRP_2 domain of transient receptor potential channels (8;17). In addition, we have also identified a chaperone binding domain in inositol hexakisphosphate kinase-2 (27), and lipid-binding domains in a host of other proteins (8;14–16). Therefore, our long-term goals are towards creating a completely automated and high-performance algorithm that contains profile sets to generate SAPPs for any known biological function. We expect that the data generated from such a tool would rapidly increase the available functional annotation for any proteome, enhancing both computational studies and biochemical/cell-biological research.
Materials and Methods
Sequences and Phylogenetic Tree Construction
All sequences used are provided in Supplemental Table 3.
For the analysis of structurally resolved SH2 domains from the PDB library, we curated the respective species specific full length sequences from RefNCBI. For convenience and readability, we annotated them with UniProtKB identifiers in Figs. 1A and Supp. Fig. 1A. Phylogenetic Analyses were performed as previously described(28).
PIP strips
as per manufacturer’s instructions using 500–1000 ng of purified protein(15).
Lipid-binding Liposomes
performed as previously described(29). Briefly, lipid mixtures phosphatidyl-ethanolamine (29.2%), phosphatidylcholine (29.2%), phosphatidylserine (29.2%), and phosphatidyl-inositol (12.5%) (all in CHCl3) were dried down to form a thin film in a 0.5-ml minifuge tube (Beckmann) and then bath sonicated in 0.2 M sucrose, 20 mM KCl, 20 mM Hepes, pH 7.4, 0.01% azide to yield a 10× dense lipid stock. This was diluted 1:10 in dilution buffer (0.12 M NaCl, 1 mM EGTA, 0.2 mM CaCl2 (free Ca2+ concentration of approximately 50 nM), 1.5 mM MgCl2, 1 mM dithiothreitol, 5 mM KCl, 20 mM Hepes, pH 7.4, 1 mg/ml bovine serum albumin) containing 500–1000 ng of recombinant protein. Protein complexes were allowed to form by incubation at 30 °C for 5 min prior to centrifugation (100,000 × g for 30 min). After spinning, supernatants were carefully removed and the pellets retrieved by addition of an equal volume of 60 °C SDS sample buffer and subsequent bath sonication. Both PIP-strip and liposomal assays were visualized via Western analysis. Films were scanned and analyzed using Bio-rad Gel-dock© system (p-values from student t-test).
Protein Purification
Mouse Tec fragments were cloned into Pet28c HIS-tagged vectors and transformed into BL21 bacteria. Protein production was induced by 100 μM IPTG for 30 minutes at 37°C. Cells were lysed by sonication in lysis buffer (PBS containing 100 mM EDTA, 1 mM PMSF, 5 mM DTT, and complete protease inhibitor mixture). After lysis, debris is pelleted by a 5 minute 10,000 × g centrifugation. The supernatant was incubated on Talon© beads for 30 minutes, washed 10 times with TBST, and eluted with 500 μl of 10 mM EDTA in TBS pH 8. Proteins were then dialyzed 2X with 4L of TBS pH 7.4 to remove detergent and EDTA.
GDDA-BLAST Boundaries
In order to generate histograms from our phylogenetic profiles which reflect structural boundaries and putative function, the profiles of related structure or function are curated. In this case we used 131 putative lipid binding domains of various length, structure, and lipid-specificity (see Supplemental Table 4 for a complete description). Each of the query sequences is compared to the chosen profiles using GDDA-BLAST. The distribution of the number of hits, above a predetermined threshold and summed over all the profiles is determined at each position of the query sequence for all of these alignments. We normalize the data by subtracting the mean number of hits per amino acid, with the histograms showing only the positive scoring regions (See Supplemental Table 1 for examples).
GDDA-BLAST Positional Analysis
by using Smith-Waterman algorithm with both settings, such as (BLOSUM62, GOP=10, GEP=0.5) and (BLOSUM45, GOP=11, GEP=1) to generate an alignment in the query sequence with profiles which were positive (>/= 60% coverage including the “seed” and >/= 10% Identity excluding the “seed”) by GDDA-BLAST. Raw scores for each residue are calculated by scoring a value=2 for identities and value=1 for positive substitutions in the alignments. These positions were tallied and the cumulative score was annotated versus the amino acid position. Following, the score was normalized for each position using the following equation.
Monte-Carlo Randomization Analysis
The human BTK sequence was randomized 10,000 times. Following, we measured multiple regions corresponding to the PH domain (a.a. 1-110), the SH3-SH2 domain (a.a. 163-392), and the kinase domain (a.a. 397-652) for the overlapping PLB domains, and normalized the score by the average # of profiles over the region measured (as performed for identifying boundaries using GDDA-BLAST).
Supplementary Material
Acknowledgments
We thank Jason Holmes at the Pennsylvania State University CAC center for technical assistance; Drs. Robert E Rothe, Jim White, Jes T. Kendall, Seth Rogan, and T.D. Dank for creative dialogue. This work was supported by the Searle Young Investigators Award, start-up monies from Pennsylvania State University (R.L.P.), the NIH R01GM087410-01 (R.L.P. and D.V.R) , the NSF 0843282 (R.L.P and D.V.R.), funds from the Huck Life Science Institute’s Center for Computational Proteomics (R.L.P and D.V.R) and a grant from the Pennsylvania Department of Health using Tobacco Settlement Funds to D.V.R. The Pennsylvania Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
References
- 1.Cai YD, Liu XJ, Xu XB, Chou KC. Support vector machines for prediction of protein domain structural class. J Theor Biol. 2003;221:115–120. doi: 10.1006/jtbi.2003.3179. [DOI] [PubMed] [Google Scholar]
- 2.Caraveo G, van Rossum DB, Patterson RL, Snyder SH, Desiderio S. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science. 2006;314:122–125. doi: 10.1126/science.1127815. [DOI] [PubMed] [Google Scholar]
- 3.Chakraborty A, Koldobskiy MA, Sixt KM, Juluri KR, Mustafa AK, et al. HSP90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang GS, Hong Y, Ko KD, Bhardwaj G, Holmes EC, et al. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc Natl Acad Sci USA. 2008;105:13474–13479. doi: 10.1073/pnas.0803860105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cokus S, Mizutani S, Pellegrini M. An improved method for identifying functionally linked proteins using phylogenetic profiles. BMC Bioinformatics. 2007;4:S7. doi: 10.1186/1471-2105-8-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, et al. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J Biol Chem. 2000;275:28261–28268. doi: 10.1074/jbc.M000469200. [DOI] [PubMed] [Google Scholar]
- 7.Grundy WN, Bailey TL. Family pairwise search with embedded motif models. Bioinformatics. 1999;15:463–470. doi: 10.1093/bioinformatics/15.6.463. [DOI] [PubMed] [Google Scholar]
- 8.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S, Henikoff JG. Embedding strategies for effective use of information from multiple sequence alignments. Protein Sci. 1997;6:698–705. doi: 10.1002/pro.5560060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Subramaniam S. Locally defined protein phylogenetic profiles reveal previously missed protein interactions and functional relationships. Proteins. 2006;62:1115–1124. doi: 10.1002/prot.20830. [DOI] [PubMed] [Google Scholar]
- 11.Ko KD, Hong Y, Chang GS, Bhardwaj G, van Rossum DB, et al. Phylogenetic Profiles as a Unified Framework for Measuring Protein Structure, Function and Evolution. Physics Archives. 2008 arXiv:0806.239, q-bio.Q. [Google Scholar]
- 12.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, et al. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin HH, Han LY, Zhang HL, Zheng CJ, Xie B, et al. Prediction of the functional class of lipid binding proteins from sequence-derived properties irrespective of sequence similarity. J Lipid Res. 2006;47:824–831. doi: 10.1194/jlr.M500530-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Machida K, Mayer BJ. The SH2 domain: versatile signaling module and pharmaceutical target. Biochim Biophys Acta. 2005;1747:1–25. doi: 10.1016/j.bbapap.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 15.Marchler BA, Anderson JB, Cherukuri PF, Weese SC, Geer LY, et al. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70. doi: 10.1007/978-1-59745-515-2_5. [DOI] [PubMed] [Google Scholar]
- 17.Mustafa AK, van Rossum DB, Patterson RL, Maag D, Ehmsen JT, et al. Glutamatergic regulation of serine racemase via reversal of PIP2 inhibition. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0813105106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikolaidis N, Chalkia D, Watkins DN, Barrow RK, Snyder SH, et al. Ancient Origin of the New Developmental Superfamily DANGER. PLoS ONE. 2007;2:e204. doi: 10.1371/journal.pone.0000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez de DR, Lopez GE, Rivera J, Ferreira A, Fontan G, et al. Naturally occurring Bruton’s tyrosine kinase mutations have no dominant negative effect in an X-linked agammaglobulinaemia cellular model. Clin Exp Immunol. 2008;152:33–38. doi: 10.1111/j.1365-2249.2008.03589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rameh LE, Chen CS, Cantley LC. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell. 1995;83:821–830. doi: 10.1016/0092-8674(95)90195-7. [DOI] [PubMed] [Google Scholar]
- 22.Ranea JA, Yeats C, Grant A, Orengo CA. Predicting protein function with hierarchical phylogenetic profiles: the Gene3D Phylo-Tuner method applied to eukaryotic genomes. PLoS Comput Biol. 2007;3:e237. doi: 10.1371/journal.pcbi.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartzberg PL, Finkelstein LD, Readinger JA. TEC-family kinases: regulators of T-helper-cell differentiation. Nat Rev Immunol. 2005;5:284–295. doi: 10.1038/nri1591. [DOI] [PubMed] [Google Scholar]
- 24.Smith TF, Waterman MS. Overlapping genes and information theory. J Theor Biol. 1981;91:379–380. doi: 10.1016/0022-5193(81)90239-3. [DOI] [PubMed] [Google Scholar]
- 25.Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 26.Tokonzaba E, Capelluto DG, Kutateladze TG, Overduin M. Phosphoinositide, phosphopeptide and pyridone interactions of the Abl SH2 domain. Chem Biol Drug Des. 2006;67:230–237. doi: 10.1111/j.1747-0285.2006.00361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Valiaho J, Smith CI, Vihinen M. BTKbase: the mutation database for X-linked agammaglobulinemia. Hum Mutat. 2006;27:1209–1217. doi: 10.1002/humu.20410. [DOI] [PubMed] [Google Scholar]
- 28.van Rossum DB, Oberdick D, Rbaibi Y, Bhardwaj G, Barrow RK, et al. TRP_2, a Lipid/Trafficking Domain That Mediates Diacylglycerol-induced Vesicle Fusion. J Biol Chem. 2008;283:34384–34392. doi: 10.1074/jbc.M804707200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rossum DB, Patterson RL, Sharma S, Barrow RK, Kornberg M, et al. Phospholipase Cgamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature. 2005;434:99–104. doi: 10.1038/nature03340. [DOI] [PubMed] [Google Scholar]
- 30.Zhu H, Bilgin M, Bangham R, Hall D, Casamayor A, et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 31.Dubchak I, Muchnik I, Holbrook SR, Kim SH. Prediction of protein folding class using global description of amino acid sequence. Proc Natl Acad Sci USA. 1995 Sep 12;92(19):8700–4. doi: 10.1073/pnas.92.19.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.