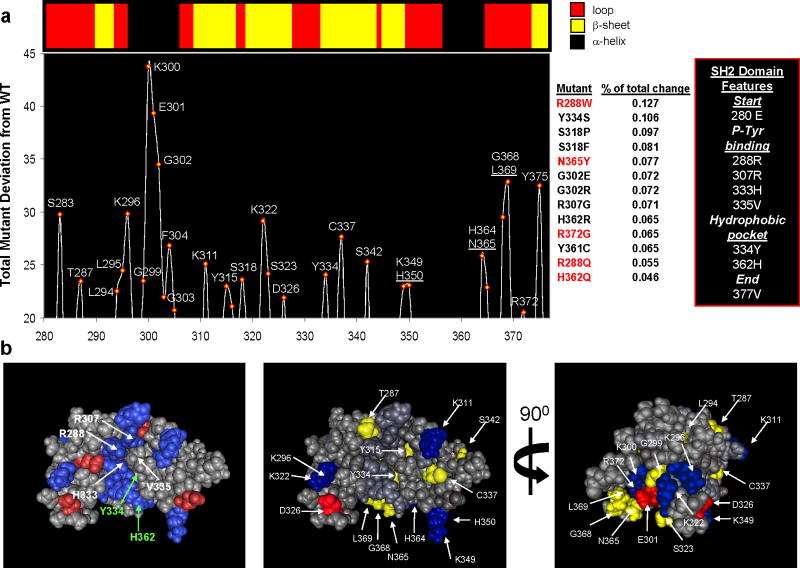
Figure 2. Phylogenetic profiles reveal putative lipid-binding residues in BTK.
(a) Top: Depiction of the secondary structural elements in the SH2 domain of human Bruton’s Tyrosine Kinase (PDB: 2GE9). Bottom: Positional analysis of human BTK for residues implicated in peripheral lipid-binding (PLB) from our phylogenetic profiles. The graph depicts the total change between sequences mutated to resemble 13 naturally occurring polymorphisms linked with XLA and wild-type sequence at each amino acid position (mutants in red were experimentally tested). Each sequence was compared with WT BTK and the absolute change recorded and finally summed for all mutations. The results indicate that the region proximal to the second phosphotyrosine-binding site (R307) is the site with the most change; however, positions throughout the p-Tyr and hydrophobic pocket can be identified. These predictions resemble those findings by Tokonzaba et al. in which they measured positions in the SH2 domain of c-Abl that display NMR structural perturbations in the presence of lipids (18)(see Supplemental Figure 1). (b) 3-D views of the SH2 domain of human Bruton’s Tyrosine Kinase (PDB: 2GE9). Left panel is colored for charged residues and labeled for those residues which make up the P-Tyrosine and Hydrophobic binding pockets. The middle and right panels are two views colored with those positions identified by GDDA-BLAST analysis. Residues colored in blue(−) and red(+) are charged.