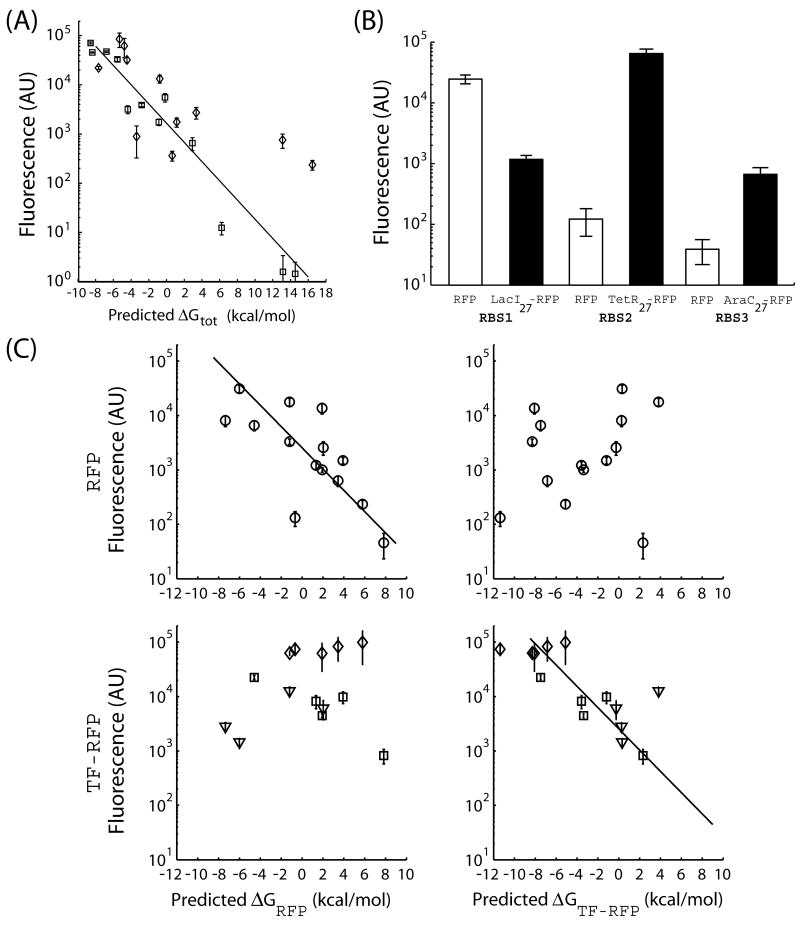
Figure 3.
The design method can control the expression level of different proteins by predicting the impact of changing the protein coding sequence. (A) The fluorescence levels from 23 synthetic RBSs in front of two different protein coding sequences are measured and compared to the predicted ΔGtot calculations. The two proteins are TetR27-RFP (diamonds) and AraC27-RFP (squares). The expected relationship between the log protein fluorescence and the predicted ΔGtot is obtained for each protein coding sequence (TetR27-RFP, R2=0.54; AraC27-RFP, R2 = 0.95). (B) Reusing the same RBS sequence with two different protein coding sequences can alter the translation initiation. Fluorescence levels from identical RBS sequences in front of either RFP (white bars) or a chimeric fluorescent protein (either LacI27-RFP, TetR27-RFP, or AraC27-RFP; black bars) are shown. (C) The design method must use the correct protein coding sequence to accurately predict the ΔGtot. The fluorescence levels from 14 pairs of RBS sequences in front of either RFP (black circles) or a chimeric fluorescent protein (LacI27-RFP, triangles; TetR27-RFP, diamonds; AraC27-RFP, squares) are measured. When the correct protein coding sequence is used to calculate the ΔGtot, the expected relationship between log protein fluorescence and ΔGtot is obtained (lines, R2 = 0.62 and R2 = 0.51). Otherwise, the thermodynamic model does not correctly predict the expression level (R2 = 0.04 and 0.02). The error bars calculated as the standard deviation of at least 6 measurements performed on 2 different days.