Table 1.
Correlation between RMSD and Zscores in MOULDER.
| Target | 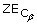 |
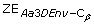 |
 |
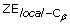 |
ZEmin | ZEAa3DEnv-min | ZE3DEnv-min | ZElocal-min |
|---|---|---|---|---|---|---|---|---|
| 1bbh | 0.86 | 0.36 | -0.83 | -0.80 | 0.62 | 0.42 | -0.02 | -0.71 |
| 1c2r | 0.71 | 0.45 | -0.48 | -0.67 | 0.69 | 0.68 | -0.43 | -0.27 |
| 1cau | 0.83 | 0.56 | -0.71 | -0.72 | 0.69 | 0.38 | -0.40 | -0.74 |
| 1cew | 0.70 | 0.31 | -0.64 | -0.58 | 0.61 | 0.08 | -0.18 | -0.63 |
| 1cid | 0.41 | -0.12 | -0.22 | -0.59 | 0.45 | 0.43 | 0.10 | -0.55 |
| 1dxt | 0.87 | 0.75 | -0.84 | -0.76 | 0.78 | 0.76 | -0.51 | -0.51 |
| 1eaf | 0.79 | 0.57 | -0.61 | -0.64 | 0.72 | 0.66 | -0.38 | -0.55 |
| 1gky | 0.88 | 0.54 | -0.77 | -0.78 | 0.73 | 0.61 | -0.18 | -0.63 |
| 1lga | 0.88 | 0.49 | -0.68 | -0.85 | 0.84 | 0.68 | -0.40 | -0.84 |
| 1mdc | 0.78 | 0.50 | -0.51 | -0.64 | 0.68 | 0.40 | -0.20 | -0.60 |
| 1mup | 0.85 | 0.66 | -0.80 | -0.83 | 0.80 | 0.79 | 0.24 | -0.79 |
| 1onc | 0.78 | 0.66 | -0.58 | -0.58 | 0.80 | 0.75 | -0.29 | -0.56 |
| 2afn | 0.86 | 0.61 | -0.60 | -0.83 | 0.77 | 0.68 | -0.27 | -0.83 |
| 2cmd | 0.81 | 0.68 | -0.82 | -0.78 | 0.63 | 0.58 | -0.46 | -0.65 |
| 2fbj | 0.77 | 0.35 | -0.32 | -0.82 | 0.79 | 0.58 | -0.13 | -0.83 |
| 2mta | 0.72 | 0.47 | -0.16 | -0.69 | 0.77 | 0.67 | -0.02 | -0.70 |
| 2pna | 0.83 | 0.58 | -0.79 | -0.55 | 0.62 | 0.52 | -0.29 | -0.46 |
| 2sim | 0.81 | -0.12 | -0.73 | -0.81 | 0.66 | 0.20 | -0.36 | -0.76 |
| 4sbv | 0.69 | -0.10 | -0.65 | -0.60 | 0.54 | 0.46 | -0.07 | -0.45 |
| 8i1b | 0.77 | 0.68 | -0.36 | -0.60 | 0.57 | 0.67 | 0.13 | -0.43 |
Pearson product-correlation between Root Mean Square Deviation (RMSD) of MOULDER decoys of 20 target/model sets (in rows) and Zscores (in columns): 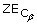 ,
, 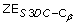 ,
, 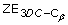 ,
, 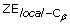 , ZEmin, ZES3DC-min, ZE3DC-min, and ZElocal-min.
, ZEmin, ZES3DC-min, ZE3DC-min, and ZElocal-min.
