Table 2.
Ranking position of the native structure according to the scoring functions.
| Target set | DOPE | GA341 | Prosa2003 | DFIRE | 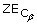 |
ZEmin |
|---|---|---|---|---|---|---|
| fisa_casp3 | ||||||
| smd3 | 1 | 51 | 2 | 1 | 12 | 1 |
| 1bg8-A | 1 | 808 | 151 | 1 | 341 | 2 |
| 1jwe | 1 | 135 | 4 | 1 | 514 | 1 |
| 1eh2 | 8 | 826 | 93 | 1 | 577 | 159 |
| 1bl0 | 1 | 809 | 729 | 1 | 458 | 3 |
| Total | 4 | 0 | 0 | 5 | 0 | 2 |
| lmds | ||||||
| smd3 | 1 | 15 | 1 | 1 | 1 | 1 |
| 2ovo | 7 | 33 | 1 | 61 | 115 | 7 |
| 1dtk | 1 | 1 | 1 | 1 | 77 | 33 |
| 4pti | 6 | 7 | 1 | 24 | 25 | 10 |
| 1b0n-B | 293 | 35 | 1 | 418 | 99 | 180 |
| 1bba | 501 | 395 | 458 | 501 | 389 | 63 |
| 1shf-A | 1 | 5 | 13 | 1 | 199 | 1 |
| 1ctf | 1 | 1 | 1 | 1 | 1 | 1 |
| 1fc2 | 501 | 234 | 107 | 501 | 276 | 489 |
| 1igd | 1 | 1 | 1 | 18 | 10 | 1 |
| 2cro | 1 | 10 | 1 | 1 | 43 | 16 |
| Total | 6 | 3 | 8 | 5 | 2 | 4 |
| 4state_reduced | ||||||
| 4rxn | 1 | 1 | 8 | 1 | 20 | 26 |
| 4pti | 1 | 6 | 1 | 1 | 1 | 1 |
| 1ctf | 1 | 1 | 1 | 1 | 1 | 1 |
| 3icb | 3 | 5 | 1 | 5 | 10 | 9 |
| 1sn3 | 1 | 1 | 1 | 1 | 25 | 3 |
| 2cro | 1 | 5 | 1 | 1 | 1 | 2 |
| 1r69 | 1 | 4 | 1 | 1 | 1 | 1 |
| Total | 6 | 3 | 6 | 6 | 4 | 3 |
| MOULDER | ||||||
| 1onc | 1 | 1 | 1 | 1 | 1 | 1 |
| 1dxt | 1 | 1 | 1 | 1 | 3 | 1 |
| 1eaf | 1 | 1 | 1 | 1 | 1 | 1 |
| 1lga | 1 | 1 | 1 | 1 | 1 | 1 |
| 1gky | 1 | 1 | 1 | 1 | 1 | 1 |
| 1cau | 1 | 1 | 1 | 1 | 1 | 1 |
| 4sbv | 1 | 1 | 1 | 1 | 1 | 1 |
| 8i1b | 1 | 1 | 1 | 1 | 1 | 1 |
| 2mta | 1 | 1 | 1 | 1 | 4 | 4 |
| 2fbj | 1 | 1 | 1 | 1 | 1 | 1 |
| 2cmd | 1 | 1 | 1 | 1 | 1 | 1 |
| 1cew | 1 | 1 | 1 | 1 | 1 | 1 |
| 2afn | 1 | 1 | 1 | 1 | 1 | 1 |
| 2sim | 1 | 1 | 1 | 1 | 1 | 1 |
| 1bbh | 1 | 1 | 1 | 1 | 1 | 1 |
| 1mdc | 1 | 1 | 1 | 1 | 1 | 1 |
| 1mup | 1 | 1 | 1 | 1 | 15 | 1 |
| 2pna | 85 | 45 | 43 | 85 | 58 | 9 |
| 1cid | 1 | 1 | 1 | 1 | 1 | 1 |
| 1c2r | 1 | 1 | 1 | 1 | 6 | 1 |
| Total | 19 | 19 | 19 | 19 | 15 | 18 |
Ranking position of the native structure among the sets of model/target decoys for several scoring functions. In the first column it is shown the code of the target protein used to generate the set of decoys. Next columns show the results for DOPE, GA341, Prosa2003, DFIRE, 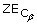 , ZEmin scoring functions. The set of decoys is split in groups: MOULDER, 4state_reduced, fisa_casp3, and lmds.
, ZEmin scoring functions. The set of decoys is split in groups: MOULDER, 4state_reduced, fisa_casp3, and lmds.
