Table 6.
Correlation between RMSD and residue-position Zscores
| Target Set | 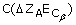 |
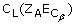 |
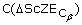 |
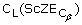 |
C(ΔZAEmin) | CL(ZAEmin) | C(ΔScZEmin) | CL(ScZEmin) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Average | P/N | Average | P/N | Average | P/N | Average | P/N | Average | P/N | Average | P/N | Average | P/N | Average | P/N | |
| 1bbh | 0.7 ± 0.1 | 124/34 | 0.7 ± 0.1 | 82/76 | 0.7 ± 0.1 | 135/23 | 0.7 ± 0.1 | 109/49 | 0.7 ± 0.1 | 112/46 | 0.6 ± 0.1 | 37/121 | 0.6 ± 0.1 | 92/66 | 0.7 ± 0.1 | 56/102 |
| 1c2r | 0.7 ± 0.1 | 87/22 | 0.6 ± 0.2 | 25/84 | 0.7 ± 0.1 | 62/47 | 0.6 ± 0.2 | 23/36 | 0.5 ± 0.2 | 10/99 | 0.6 ± 0.2 | 13/96 | 0.5 ± 0.2 | 6/103 | 0.5 ± 0.2 | 5/104 |
| 1cau | 0.6 ± 0.2 | 22/16 | 0.7 ± 0.2 | 28/10 | 0.5 ± 0.2 | 4/34 | 0.7 ± 0.2 | 28/10 | 0.6 ± 0.2 | 11/27 | 0.7 ± 0.2 | 26/12 | 0.5 ± 0.2 | 7/31 | 0.6 ± 0.2 | 19/19 |
| 1cew | 0.8 ± 0.0 | 1/11 | 0.5 ± 0.3 | 3/9 | 0.6 ± 0.0 | 1/11 | 0.8 ± 0.0 | 1/11 | 0.6 ± 0 | 1/11 | 0.5 ± 0.4 | 2/10 | 0.0 ± 0.0 | 0/12 | 0.8 ± 0.0 | 1/11 |
| 1cid | 0.7 ± 0.1 | 56/27 | 0.8 ± 0.2 | 52/31 | 0.7 ± 0.1 | 60/23 | 0.7 ± 0.1 | 59/24 | 0.8 ± 0.1 | 77/6 | 0.7 ± 0.2 | 28/55 | 0.8 ± 0.1 | 80/3 | 0.7 ± 0.2 | 28/55 |
| 1dxt | 0.6 ± 0.1 | 44/106 | 0.7 ± 0.1 | 89/61 | 0.6 ± 0.1 | 79/71 | 0.7 ± 0.1 | 111/39 | 0.5 ± 0.2 | 6/144 | 0.7 ± 0.2 | 46/104 | 0.5 ± 0.2 | 8/142 | 0.7 ± 0.1 | 56/94 |
| 1eaf | 0.5 ± 0.2 | 10/50 | 0.6 ± 0.2 | 18/42 | 0.6 ± 0.2 | 15/45 | 0.5 ± 0.1 | 22/38 | 0.4 ± 0.2 | 3/57 | 0.6 ± 0.2 | 17/43 | 0.4 ± 0.2 | 3/57 | 0.6 ± 0.1 | 28/32 |
| 1gky | 0.6 ± 0.2 | 12/17 | 0.7 ± 0.2 | 14/5 | 0.5 ± 0.2 | 5/14 | 0.6 ± 0.2 | 14/5 | 0.6 ± 0.2 | 13/6 | 0.6 ± 0.2 | 10/9 | 0.6 ± 0.2 | 13/6 | 0.7 ± 0.2 | 12/7 |
| 1lga | 0.5 ± 0.2 | 12/95 | 0.5 ± 0.2 | 10/97 | 0.5 ± 0.2 | 9/98 | 0.5 ± 0.2 | 5/102 | 0.6 ± 0.0 | 1/106 | 0.5 ± 0.2 | 8/99 | 0.4 ± 0.3 | 2/105 | 0.4 ± 0.3 | 3/104 |
| 1mdc | 0.7 ± 0.1 | 59/56 | 0.7 ± 0.2 | 42/73 | 0.7 ± 0.1 | 68/47 | 0.7 ± 0.2 | 47/68 | 0.6 ± 0.1 | 39/76 | 0.6 ± 0.1 | 28/87 | 0.6 ± 0.2 | 16/99 | 0.6 ± 0.2 | 18/97 |
| 1mup | 0.7 ± 0.1 | 60/74 | 0.7 ± 0.1 | 73/61 | 0.7 ± 0.1 | 68/66 | 0.7 ± 0.1 | 77/57 | 0.7 ± 0.1 | 99/35 | 0.7 ± 0.1 | 59/75 | 0.7 ± 0.1 | 112/22 | 0.7 ± 0.1 | 53/81 |
| 1onc | 0.7 ± 0.2 | 53/69 | 0.6 ± 0.2 | 29/93 | 0.7 ± 0.2 | 59/63 | 0.7 ± 0.2 | 38/84 | 0.7 ± 0.1 | 102/20 | 0.7 ± 0.2 | 43/79 | 0.7 ± 0.1 | 86/36 | 0.7 ± 0.1 | 40/82 |
| 2afn | 0.6 ± 0.1 | 80/39 | 0.6 ± 0.1 | 73/46 | 0.6 ± 0.1 | 22/97 | 0.6 ± 0.1 | 71/48 | 0.5 ± 0.1 | 25/94 | 0.5 ± 0.2 | 18/103 | 0.5 ± 0.2 | 9/110 | 0.5 ± 0.1 | 17/102 |
| 2cmd | 0.7 ± 0.1 | 101/128 | 0.6 ± 0.1 | 112/117 | 0.6 ± 0.1 | 93/136 | 0.6 ± 0.1 | 103/126 | 0.6 ± 0.1 | 102/127 | 0.6 ± 0.1 | 50/179 | 0.6 ± 0.1 | 42/187 | 0.6 ± 0.1 | 58/171 |
| 2fbj | 0.6 ± 0.2 | 12/89 | 0.6 ± 0.1 | 24/77 | 0.6 ± 0.2 | 7/94 | 0.6 ± 0.2 | 13/88 | 0.6 ± 0.1 | 32/69 | 0.6 ± 0.2 | 11/90 | 0.6 ± 0.1 | 40/61 | 0.6 ± 0.1 | 19/82 |
| 2mta | 0.6 ± 0.1 | 47/111 | 0.7 ± 0.1 | 48/110 | 0.7 ± 0.1 | 75/83 | 0.7 ± 0.2 | 32/126 | 0.7 ± 0.1 | 104/54 | 0.7 ± 0.1 | 73/85 | 0.7 ± 0.1 | 130/28 | 0.6 ± 0.2 | 40/118 |
| 2pna | 0.7 ± 0.1 | 41/97 | 0.7 ± 0.1 | 73/65 | 0.7 ± 0.2 | 37/101 | 0.7 ± 0.1 | 71/67 | 0.0 ± 0.0 | 0/138 | 0.7 ± 0.2 | 67/71 | 0.6 ± 0.2 | 12/126 | 0.7 ± 0.2 | 59/79 |
| 2sim | 0.5 ± 0.2 | 4/90 | 0.4 ± 0.3 | 3/91 | 0.0 ± 0.0 | 0/94 | 0.6 ± 0.0 | 1/93 | 0.4 ± 0.3 | 2/92 | 0.6 ± 0.0 | 1/93 | 0.4 ± 0.3 | 2/92 | 0.0 ± 0.0 | 0/94 |
| 4sbv | 0.5 ± 0.4 | 2/2 | 0.4 ± 0.3 | 2/2 | 0.4 ± 0.3 | 2/2 | 0.4 ± 0.3 | 2/2 | 0.0 ± 0.0 | 0/4 | 0.0 ± 0.0 | 0/4 | 0.0 ± 0.0 | 0/4 | 0.0 ± 0.0 | 0/4 |
| 8i1b | 0.4 ± 0.3 | 2/135 | 0.6 ± 0.2 | 11/126 | 0.5 ± 0.2 | 5/132 | 0.6 ± 0.2 | 13/124 | 0.6 ± 0.1 | 43/94 | 0.5 ± 0.1 | 18/119 | 0.5 ± 0.1 | 23/114 | 0.6 ± 0.2 | 13/124 |
Pearson correlation between RMSD of Cα atoms and residue-position Zscores of structure-models in MOULDER decoy set. In the first column is shown the code of the native protein used to generate the decoys of a model/target set. Next columns show: i) the average of Pearson correlation (Average) of those models with RMSD from the native structure smaller than 7Å and using only correlations higher than 0.5; and ii) the ratio P/N, being P the number of models with correlation larger than 0.5 and N those with correlation smaller than or equal to 0.5 among models with RMSD larger than 7Å. Residue-position Zscores are: Sc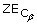 , ScZEmin,
, ScZEmin, 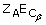 , ZAEmin and the differences of Sc
, ZAEmin and the differences of Sc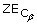 and ScZEmin of the decoy conformers with respect to their native structure (ΔSc
and ScZEmin of the decoy conformers with respect to their native structure (ΔSc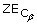 and ΔScZEmin). Pearson correlations between Cα RMSDs and Zscores are denoted as C(Zscore) - in even columns -, while correlation of Cα RMSDs and Zscores normalized by length are indicated as CL(Zscore) - in odd columns -.
and ΔScZEmin). Pearson correlations between Cα RMSDs and Zscores are denoted as C(Zscore) - in even columns -, while correlation of Cα RMSDs and Zscores normalized by length are indicated as CL(Zscore) - in odd columns -.
