Abstract
Cystathionine β-synthase (CBS) is a homocysteine metabolizing enzyme that contains pyridoxal phosphate (PLP) and a six-coordinate heme cofactor of unknown function. CBS was inactivated by peroxynitrite, the product of nitric oxide and superoxide radicals. The IC50 was ~150 μM for 5 μM ferric CBS. Stopped-flow kinetics and competition experiments showed a direct reaction with a second-order rate constant of (2.4–5.0) × 104 M−1 s−1 (pH 7.4, 37 °C). The radicals derived from peroxynitrite, nitrogen dioxide and carbonate radical, also inactivated CBS. Exposure to peroxynitrite did not modify bound PLP but led to nitration of Trp208, Trp43 and Tyr223 and alterations in the heme environment including loss of thiolate coordination, conversion to high spin and bleaching, with no detectable formation of oxo-ferryl compounds nor promotion of one-electron processes. This study demonstrates the susceptibility of CBS to reactive oxygen/nitrogen species, with potential relevance to hyperhomocysteinemia, a risk factor for cardiovascular diseases.
Keywords: Cystathionine β-synthase, Heme, Pyridoxal 5′-phosphate, Peroxynitrite, Nitrotyrosine, Nitrotryptophan, Homocysteine, Hyperhomocysteinemia
Introduction
Mammalian cystathionine β-synthase (CBS, E.C. 4.2.1.22)1 catalyzes the condensation of homocysteine with serine to form cystathionine and water in the first step of the transsulfuration pathway that yields cysteine. CBS can also catalyze the formation of hydrogen sulfide, a novel gasotransmitter with signaling and cytoprotective effects [1].
Homocysteine is a toxic metabolite that is elevated in hyperhomocysteinemic patients and constitutes an independent risk factor for cardiovascular diseases, including atherosclerosis. Additionally, elevations in plasma homocysteine correlate with other pathologies such as neurological disorders [2–4]. The most common cause of inherited hyperhomocysteinemia is deficiency in CBS. Presently, 140 mutations have been described and are listed at http://cbs.lf1.cuni.cz/index.php.
The human full-length CBS is a homotetramer of 63 kDa subunits that exhibit a modular organization [5, 6]. Each subunit contains at the N-terminal end the binding site for a heme b-type cofactor. The middle catalytic core has a pyridoxal 5′-phosphate (PLP) cofactor essential for catalysis and a CXXC disulfide oxidoreductase motif whose function has not been determined. Finally, the C-terminal region contains a tandem repeat of two “CBS domains” that probably bind S-adenosyl-L-methionine (SAM), an allosteric activator [7]. The regulatory region can be cleaved proteolytically by trypsin, yielding a truncated dimeric enzyme with 45 kDa subunits that is more active, stable, and unresponsive to SAM [5]. This truncated form can be observed in liver cells exposed to the proinflammatory cytokine, tumoral necrosis factor [8].
CBS is the only known PLP-dependent enzyme that also contains heme [9]. The heme cofactor is six-coordinate and in the low spin state with cysteine (Cys52, human numbering) and histidine (His65) as axial ligands [10–12]. The UV-visible absorption spectrum of ferric CBS (Fe(III)CBS) shows a δ peak at 364 nm, a Soret peak at 428 nm and a broad absorption feature at 550 nm corresponding to the αβ bands [13]. The coordination between the thiolate and ferric iron can be observed at high enzyme concentrations (~100 μM) in the low energy region by the presence of two bands at 645 and 705 nm [14]. Upon reduction to Fe(II)CBS with sodium dithionite or titanium citrate, the Soret peak shifts to 449 nm and the αβ bands are resolved into two peaks at 571 and 540 nm [13].
The function of the heme is yet unknown. The six-coordinated hemethiolate has very low reactivity in the ferric state and this stability is reflected by the fairly low reduction potential of −0.291 V for the truncated enzyme [15]. It is unreactive towards typical ferric heme ligands like cyanide, fluoride, azide, pyridines, amines, isonitriles and imidazoles [16, 17]. However, treatment of the ferric enzyme with high concentrations of the thiol chelator mercuric chloride (HgCl2) results in the conversion of the six-coordinate low-spin heme to a five-coordinate high-spin species, which is paralleled by a loss of activity [12, 18]. Indeed, perturbations in the heme environment lead to enzyme inactivation. For example, reduction at high temperatures, which leads to loss of the native cysteinate ligand and formation of a ferrous species with a neutral ligand that absorbs at 424 nm, inactivates the enzyme [19]. Analogously, mutation of the axial ligands His65 and Cys52 decrease activity despite the relatively high PLP content of the resulting protein [20]. It has been postulated that communication between the PLP and heme domains, distant 20 Ǻ, may be mediated by an α-helix (helix 8) that interacts at one end with the Cys52 ligand via Arg266, and at the other end with PLP [21, 22].
Peroxynitrite2 (ONOO−) is a powerful oxidizing and nitrating agent that is produced in biological systems by the reaction of superoxide (O2•−) and nitric oxide (•NO) [23, 24]. Due to its pKA of 6.8, the anion predominates (80% at pH 7.4) under physiological conditions. The conjugated acid homolyzes at a rate of 0.9 s−1 (37 °C, pH 7.4), giving free hydroxyl (•OH) and nitrogen dioxide (•NO2) radicals in 30% yield, which can nitrate aromatic residues such as tyrosine and tryptophan. The preferential biotargets of peroxynitrite are metal centers, carbon dioxide, and sulfur and selenium compounds (for recent reviews see [25, 26]). The reaction with carbon dioxide (4.6 × 104 M−1 s−1 at 37 °C and pH 7.4) [27] leads to the formation of carbonate radical (CO3•−) and nitrogen dioxide in 35% yield.
Peroxynitrite reacts with ferric heme proteins including hemethiolates at rates ranging from 104 to 107 M−1 s−1. The reaction usually proceeds through the intermediate formation of oxo-ferryl species [28–30]. Depending on the protein, the outcome can be decomposition of peroxynitrite to nitrate and nitrite or enhancement of one-electron oxidative processes, sometimes leading to enzyme inactivation and increased nitration of exogenous or endogenous aromatic residues [29, 31, 32].
Compelling evidence shows that reactive oxygen and nitrogen species are central mediators of several pathological conditions including cardiovascular disease. The increased formation of peroxynitrite in vivo is confirmed by the observed effect of superoxide radical on nitric oxide bioavailability and by the detection of nitrotyrosine (for reviews see [33, 34]). Since peroxynitrite interaction with CBS could lead to enzyme inactivation and homocysteine accumulation, we investigated the reactivity of peroxynitrite with the human truncated Fe(III)CBS dimer in this study.
Materials and methods
Peroxynitrite synthesis
The stock solutions were prepared from acidified hydrogen peroxide and sodium nitrite in a tandem quenched-flow mixing apparatus [35]. The solutions were treated with manganese dioxide to remove residual hydrogen peroxide. The concentration of peroxynitrite was determined from its absorbance at 302 nm (ε = 1670 M−1 cm−1 [36]). The concentration of nitrite present as contaminant was measured with the Griess reagent [37] after decay of peroxynitrite to nitrate in monobasic sodium phosphate solution. Nitrite content was always less than 30% of peroxynitrite. Peroxynitrite was diluted in 0.1 M NaOH immediately before use. Stock solutions were stored at −80 °C, used only once after thawing, and then discarded.
Enzyme purification
Truncated human CBS lacking 143 amino acids at the C-terminus (CBS C143) was purified as a fusion protein with glutathione S-transferase using the E. coli expression vector pGEXCBSN [38], kindly provided by Dr. Warren Kruger, Fox Chase Cancer Center, Philadelphia. The protein was purified as described previously [13] through affinity chromatography with glutathione sepharose and anion exchange chromatography. The glutathione S-transferase tag was cleaved by limited proteolysis using thrombin. The protein obtained is an active and stable dimer of 45 kDa subunits that does not bind SAM.
Enzymatic assays
CBS activity in Tris buffer (0.1 M, pH 8.3) was determined using the ninhydrin assay [39]. The specific activity of the enzyme was ~9.8 μmol min−1 mg−1 at 37 °C, similar to previously reported [5]. The protein concentration was determined by the Bradford method using albumin as a standard. The ratio of absorbance at 280 and 428 nm was 1.1. Thiols were measured spectrophotometrically using 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, ε412 = 13600 M−1 cm−1 [40]), after ultrafiltration with Ultrafree 0.5 centrifugal filter devices (Millipore) to remove interference from the heme and PLP at 412 nm.
Exposure of CBS to peroxynitrite
Peroxynitrite was added to the enzyme in sodium phosphate buffer of the specified concentration and pH, in the presence of diethylenetriaminepentaacetic acid (DTPA, 0.1 mM) to eliminate potential metal trace interference. In order to minimize changes in pH due to NaOH present in peroxynitrite solutions, additions were always < 5% of total volume. The enzyme was incubated for 2 min at 37 °C before and after peroxynitrite was added. Reverse order of addition (ROA) controls, where peroxynitrite was decomposed in buffer before mixing with enzyme, were performed with the higher concentration of peroxynitrite used in each experiment. Phosphate buffers were prepared daily avoiding the use of NaOH in order to minimize bicarbonate/carbon dioxide contamination. For experiments in which sodium bicarbonate (25 mM) was specifically added, the initial pH of the buffer was lowered so that the final value was 7.4 ± 0.1. The buffers were prepared immediately before the experiment and used within 10 min to minimize diffusion of carbon dioxide out of the solution.
CBS reduction
Ferrous CBS was obtained by the addition of known amounts of sodium dithionite (Na2S2O4). Dithionite stock solutions were prepared in degassed 0.1 N NaOH and quantified by ferricyanide reduction (ε420 = 1020 M−1 cm−1) [41] assuming a 2:1 stoichiometry.
UV-visible spectra
Fe(III)CBS was mixed with peroxynitrite in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM). After ~2 min at 37 °C, spectra were recorded in a Varian Cary 50 spectrophotometer. In some cases, after exposure to peroxynitrite the samples were reduced with dithionite in order to obtain the spectra of Fe(II)CBS.
PLP analysis
The internal PLP aldimine was removed from Fe(III)CBS (7 μM) by incubation with hydroxylamine (5 mM) in phosphate buffer (0.1 M, pH 7.2) at 4 °C for 70 h to generate an oxime that was then separated by ultrafiltration [16]. The oxime was determined fluorimetrically at 446 nm following excitation at 353 nm in an Aminco-Bowman Series 2 luminescence spectrometer. Calibration curves were made using known concentrations of PLP and 5 mM hydroxylamine in phosphate buffer (0.1 M, pH 7.4).
Kinetic studies
The reaction of Fe(III)CBS with peroxynitrite was studied following peroxynitrite decay at 37.0 ± 0.1 °C in the absence and presence of increasing concentrations of enzyme in a stopped-flow spectrophotometer (Applied Photophysics, SF.17MV) with a mixing time of less than 2 ms. The wavelength used was 290 nm instead of 302 nm to minimize absorbance changes of CBS. The apparent rate constant was determined using an initial rate approach described previously [42, 43] where the first 0.1–0.2 s were fitted to a linear plot and the apparent rate constant was determined as −(dA/dt)/(A0−A∞), the ratio between the slope and the difference between the initial and the final absorbance. Two hundred absorbance measurements were acquired during the first 0.2 s and 200 additional points were acquired until more than 99.9% of the peroxynitrite had decomposed (0.2–10 s). The pH was measured after mixing peroxynitrite with buffer, and two independent experiments were performed with similar results. Time-dependent spectra were obtained with a photodiode array accessory.
The rate constant was confirmed by a competition assay with ferrous cytochrome c (Fe(II)cyt c), that was prepared immediately before use by reduction of Fe(III)cyt c with sodium dithionite followed by gel filtration. Fe(II)cyt c concentration was determined at 550 nm (ε550 = 21000 M−1 cm−1 [44]).
Nitration of free tyrosine
To study the effect of Fe(III)CBS in the generation of nitrating species from peroxynitrite, free tyrosine (0.5 mM) was exposed to peroxynitrite (0.5 mM) in the presence and absence of CBS (10 μM) in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM). The samples were then ultrafiltered and nitrotyrosine was quantified both by absorbance at 430 nm at pH 10.5 (ε430 = 4400 M−1 cm−1 [45]) and by HPLC using a C18 reverse phase column and an isocratic mobile phase of 6.5% acetonitrile and 0.1% trifluoroacetic acid, with UV detection.
Electron paramagnetic resonance (EPR) spectroscopy
EPR spectra were recorded on a Bruker ESP 300E spectrometer equipped with an Oxford ITC4 temperature controller, a Model 5340 automatic frequency counter from Hewlett-Packard, and a gaussmeter. The specific conditions used for spectra acquisition are described in the figure legend. The heme concentration was determined independently by UV-visible absorption spectroscopy (ε428 = 73,500 M−1 cm−1 [15]) and by spin quantitation comparing the second integral of the sample spectrum with that of a 1 mM cupric perchlorate standard and were found to be similar.
HPLC analysis of heme
An HPLC system was used to investigate heme modifications [46]. CBS (5 μM), control or treated with peroxynitrite (500 μM), was concentrated to 1 mg mL−1 by ultrafiltration with Ultrafree 0.5 centrifugal filter devices (Millipore) before HPLC injection (1 μL). Both samples were analyzed using a C4 reverse phase column (5 μm, 100 mm × 300 μm, 300 Å, Vydac MS). The solvent system consisted of solution A (0.1% trifluoroacetic acid, water) and solution B (0.07% trifluoroacetic acid, acetonitrile). The column was eluted with a linear gradient from 33% to 60% B over 60 min at a flow rate of 4 μL min−1 and each sample was monitored at 210, 280, 400 and 428 nm [46]. For each sample, the main peak at 400 nm was recovered and analyzed by matrix assisted laser desorption ionization-time of flight mass spectrometry in a 4800 MALDI TOF-TOF Analyzer (Applied Biosystems). Mass spectra were acquired in reflector mode using a matrix solution of 2,5-dihydroxybenzoic acid. In samples where heme was identified (theoretical monoisotopic m/z 616.18), MS/MS analysis was performed.
Reductive alkylation, trypsin digestion and mass spectrometry
CBS (5 μM), control or treated with peroxynitrite (500 μM), was incubated with guanidine (6 M) and dithiothreitol (252 μM) for 2 h at 37 °C, under nitrogen. Iodoacetamide (1.26 mM) was added for 1 h in the darkness. Then, β-mercaptoethanol (12.6 mM) was added and the incubation continued for 30 min. The samples were washed with ammonium bicarbonate (0.1 M, pH 8) by ultrafiltration. Trypsin (Promega) was incubated with CBS in a 1/50 molar ratio for 18 h at 37 °C. Peptides resulting from proteolytic cleavage were analyzed by MALDI TOF MS. Mass spectra were acquired for positive ions in reflector mode using a matrix solution of -cyano-4-hydroxycinnamic acid in 0.2% trifluoroacetic acid and 50% acetonitrile and were externally calibrated using a mixture of peptide standards (Applied Biosystems). Samples for MS were prepared by spotting 0.5 μL of matrix solution and 0.5 μL of sample, or tiny droplets from desalting microcolumns (Omix Tip C18MB, 10 μL, Varian Inc.) eluted with matrix solution, directly on the sample plate. The amino acid sequence information was obtained from the Swiss-Prot database (P35520). Virtual tryptic digestions were performed with the GPMAW32 (v.4.02) program (Lighthouse Data). Selected putative nitrated peptides, characterized by an increase in 45 Da with regard to native peptides, were fragmented by collision-induced dissociation (CID) MS/MS analysis on a MALDI TOF/TOF instrument. Sequence identification was performed by Swiss-Prot database searching with precursor and fragment ion m/z values, using Mascot sequence query searching engine (Matrix Science, http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=SQ). Search parameters were set as follows: taxonomy, Homo sapiens; monoisotopic mass tolerance, 0.05 Da; MS/MS tolerance, 0.2 Da; partial methionine oxidation and tryptophan nitration allowed as modifications.
Results
CBS inactivation by peroxynitrite
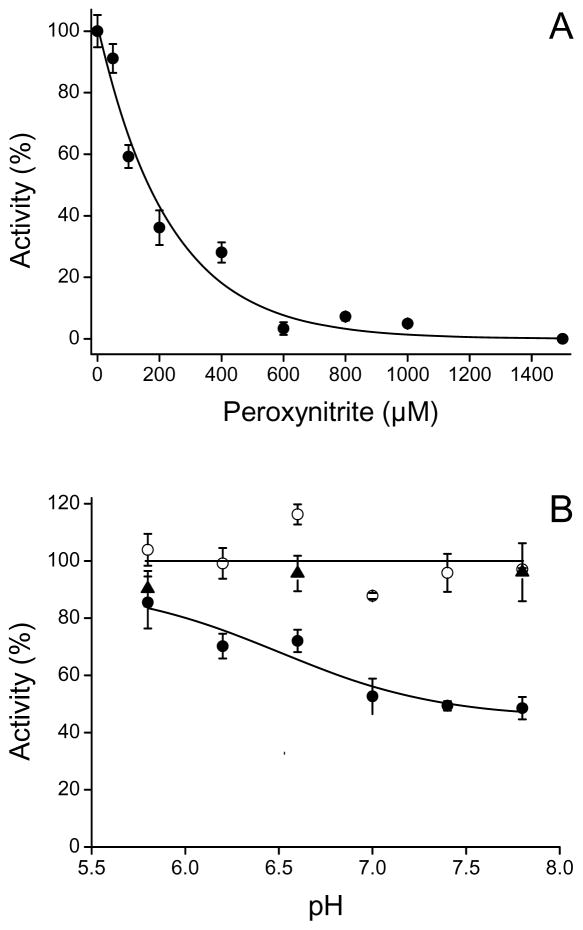
Exposure of Fe(III)CBS to peroxynitrite resulted in a dose-dependent inactivation (Fig. 1A). At concentrations above 1000 μM, total inactivation was observed. The IC50 for inactivation (concentration of peroxynitrite that inactivated by 50%) was ~150 μM for 5 μM CBS. This value is comparable to those reported for Cu, Zn superoxide dismutase [47] and tyrosine hydroxylase [48] indicating a moderate sensitivity of CBS to peroxynitrite. The inactivation was higher when Fe(III)CBS was exposed to peroxynitrite at alkaline rather than acidic pH (Fig. 1B), suggesting the involvement of peroxynitrite anion. Reverse order of addition controls ruled out a role of potential peroxynitrite impurities in the inactivation.
Fig. 1.
Enzyme inactivation with peroxynitrite. (A) Fe(III)CBS (5 μM) was incubated with increasing concentrations of peroxynitrite in phosphate buffer (pH 7.4, 0.1 M, DTPA 0.1 mM). After 2 min at 37 °C, the remaining activity was determined at pH 8.3. (B) Fe(III)CBS (5 μM) was exposed to peroxynitrite (200 μM, filled circles) in phosphate buffers of different pH. Controls without peroxynitrite (open circles) and with decomposed peroxynitrite (triangles, reverse order of addition controls) were performed. Data are the means ± standard deviations of triplicates for a representative experiment.
Kinetics of peroxynitrite reaction with CBS
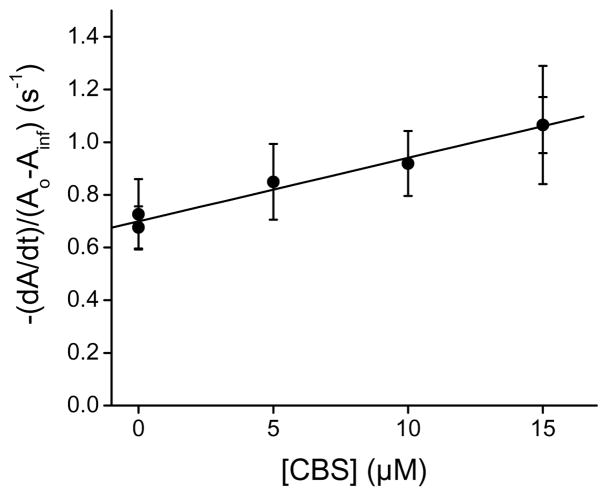
An initial rate approach was used to study the kinetics using stopped-flow spectrophotometry, because pseudo-first-order concentrations of CBS with respect to peroxynitrite could not be achieved [42, 43]. As shown in Fig. 2, the rate of peroxynitrite decay increased with the concentration of CBS, showing that a direct reaction occurs between the enzyme and peroxynitrite. From the slope of the plot, a second-order rate constant of (2.4 ± 0.3) × 104 M−1 s−1 at pH 7.4 and 37 °C was calculated.
Fig. 2.
Stopped-flow kinetics of peroxynitrite decay in the presence of CBS. Peroxynitrite (50 μM) was mixed with increasing concentrations of Fe(III)CBS in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM) at 37.0 ± 0.1 °C. The absorbance at 290 nm was recorded for 10 s and the apparent rate constant of peroxynitrite decay was determined from the initial slope. The results are the means ± standard deviation (n ≥ 10) of a representative experiment.
The direct reaction between peroxynitrite and Fe(III)CBS was confirmed by a competition assay. Fe(II)cyt c is oxidized to Fe(III)cyt c by peroxynitrite with a second-order rate constant of 2.4 × 104 M−1 s−1 (37 °C, pH 7.4) [49]. The competing reactions are described by equations 1 and 2.
| equation 1 |
| equation 2 |
In our experimental conditions, 200 μM Fe(II)cyt c and 167 μM CBS were mixed with 100 μM peroxynitrite. It can be calculated that ~90% of peroxynitrite reacted directly with the targets instead of undergoing homolysis, which occurs with a rate constant of 0.9 s−1. Equation 3 was used to calculate the second-order rate constant for the reaction of peroxynitrite with Fe(III)CBS [50], where the concentration of PCBS was calculated from the absorbance data at 550 nm as the difference in concentration of cytochrome c oxidized by peroxynitrite in the absence and presence of CBS.
| equation 3 |
The amount of Fe(II)cyt c remaining after oxidation increased in the presence of CBS from 70 μM to 148 μM, so the second-order rate constant was calculated as 5.0 × 104 M−1 s−1, similar to the results obtained by stopped-flow spectrophotometry.
Effects of mannitol, p-hydroxyphenylacetic acid (p-HPA) and carbon dioxide on the peroxynitrite-dependent inactivation of CBS
The kinetic results show that Fe(III)CBS reacts directly with peroxynitrite, but in addition the enzyme might interact with the radicals derived from peroxynitrite homolysis, nitrogen dioxide and hydroxyl radicals. In order to evaluate the incidence of these species in CBS inactivation, we exposed the enzyme to peroxynitrite in the presence of mannitol, a known hydroxyl radical scavenger, and p-HPA, a soluble tyrosine analogue that reacts with both nitrogen dioxide and hydroxyl radicals. While mannitol had no effect, p-HPA partially protected the enzyme (Table 1) suggesting that nitrogen dioxide contributes to the observed inactivation.
Table 1.
Effect of mannitol, p-HPA and bicarbonate on peroxynitrite-dependent CBS inactivation
| Condition a | CBS activity (U/mg) |
|---|---|
| CBS | 561 ± 35 b |
| + Peroxynitrite | 105 ± 1 |
| + Peroxynitrite + mannitol | 67 ± 5 |
| + Mannitol | 611 ± 47 |
| + Peroxynitrite + p-HPA | 269 ± 11 |
| + p-HPA | 521 ± 5 |
| + Peroxynitrite + NaHCO3 | 48 ± 3 |
| + NaHCO3 | 536 ± 29 |
CBS (5 μM) was exposed to peroxynitrite (400 μM) in the presence and absence of mannitol (100 μM), p-HPA (91 μM) and NaHCO3 (25 mM) in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM). After 2 min at 37 °C the enzymatic activity was determined by the ninhydrin method.
Data are the mean ± standard deviation of triplicates.
Peroxynitrite reacts directly with carbon dioxide with a second-order rate constant of 4.6 × 104 M−1 s−1 [27] yielding carbonate and nitrogen dioxide radicals. This reaction is physiologically relevant since dissolved carbon dioxide concentrations are high (1–2 mM). To investigate a possible effect of carbonate radical in CBS inactivation, we added sodium bicarbonate (25 mM) to the buffer, so that the concentration of carbon dioxide was 1.2 mM at pH 7.4 according to the pKA of 6.1 for the bicarbonate/carbon dioxide pair. This amount of carbon dioxide, which is able to trap virtually all the peroxynitrite, did not protect CBS from inactivation, suggesting that carbonate radical, in addition to nitrogen dioxide, is able to inactivate CBS.
PLP in CBS is not modified by peroxynitrite
Due to the fact that UV–visible spectral changes were observed in free PLP treated with peroxynitrite (data not shown), we studied whether the PLP bound to the enzyme could also be modified. After removal of PLP from CBS with hydroxylamine, the concentration of released oxime obtained was similar in controls (no peroxynitrite or reverse addition) and in CBS treated with 200, 500 and 800 μM peroxynitrite. Assuming that each CBS monomer has 1 molecule of PLP, we observed complete cofactor recovery under all conditions. Furthermore, control and treated CBS led to similar fluorescence spectra of the released PLP oxime, suggesting that peroxynitrite does not modify bound PLP (data not shown).
Although the fluorescence of PLP bound to CBS is difficult to interpret because of heme interference, the intrinsic fluorescence spectra of both treated and control CBS samples showed similar emission peaks at 508 nm (protonated form, λexc 420 nm) and 388 nm (deprotonated form, λexc 330 nm) [6, 16, 51]. On the other hand, when the sample was excited at 280 nm to evaluate possible fluorescence changes to the six tryptophan residues of truncated CBS, the emission spectra showed a peak at 340 nm that decreased in intensity with peroxynitrite treatment to 82% and 69% with 200 and 500 μM peroxynitrite respectively. This result suggests that tryptophan residues were modified (data not shown).
Thiol decay in CBS exposed to peroxynitrite
In native CBS, one free thiol per CBS monomer was quantified with DTNB under our experimental conditions, consistent with the reported value. This thiol can be assigned to Cys15, a residue that is non-critical for activity [52]. Exposure of Fe(III)CBS (5 μM) to increasing concentrations of peroxynitrite led to the expected decrease in thiol concentration, with an IC50 of ~50 μM (data not shown). This value is one third of the IC50 for inactivation, consistent with the measured thiol being non-critical.
Changes in the UV-visible spectra of CBS upon exposure to peroxynitrite
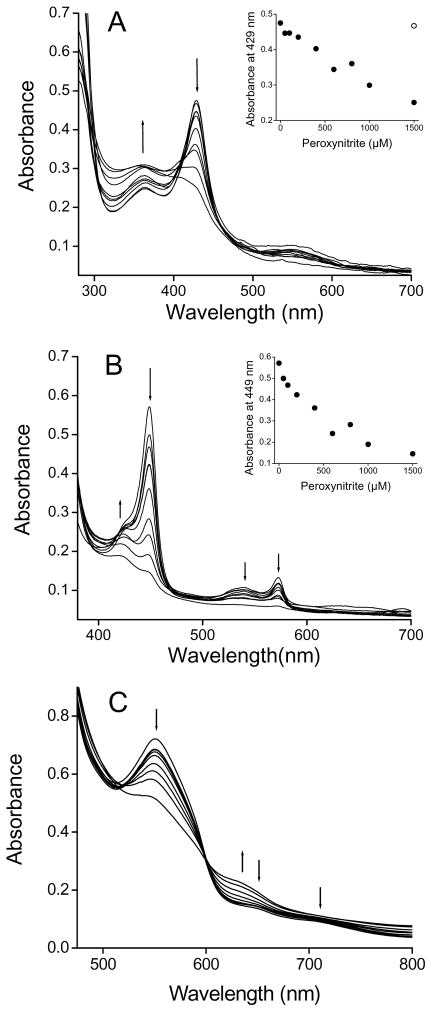
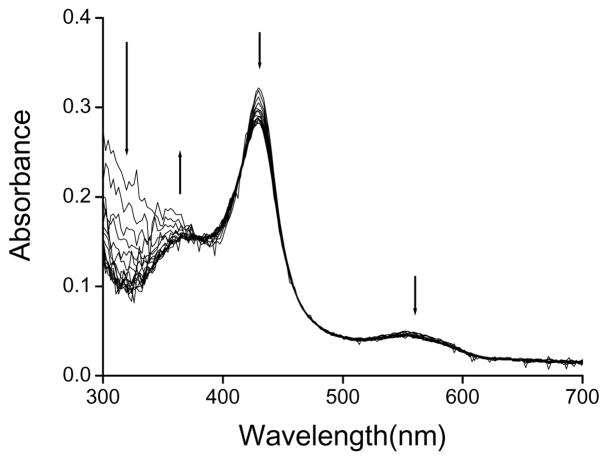
The UV-visible spectra of Fe(III)CBS treated with peroxynitrite showed a decrease in the absorbance of the Soret maximum and a blue shift from 429 to 425.5 nm together with an increase at 364 nm. At concentrations above 800 μM peroxynitrite, the typical heme spectra was dramatically modified (Fig. 3A). When peroxynitrite was added in consecutive 50 μM aliquots, the changes in the electronic absorption spectra were qualitatively similar (data not shown).
Fig. 3.
UV-visible spectra of CBS treated with peroxynitrite. (A) Samples containing Fe(III)CBS (5 μM) in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM) were mixed with increasing concentrations of peroxynitrite (0, 50, 100, 200, 400, 600, 800, 1000 and 1500 μM) and spectra were recorded after ~2 min at 37 °C. (Inset) Absorbance at 429 nm versus peroxynitrite concentration. The open symbol represents the reverse order of addition control. (B) Fe(III)CBS was exposed to peroxynitrite as in (A). Samples were then reduced to Fe(II)CBS with dithionite (3.5 mM) and the spectra recorded. (Inset) Absorbance at 449 nm versus peroxynitrite concentration. (C) Fe(III)CBS (123 μM) in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM) was mixed with consecutive additions of peroxynitrite (0, 100, 200, 300, 500, 1000, 1500, 2000 and 3000 μM, final concentrations) and spectra were recorded ~2 min after each addition.
When these samples were reduced with dithionite to Fe(II)CBS, the absorbance of the Soret peak at 449 nm decreased. At concentrations between 200 and 800 μM peroxynitrite, a peak at 424.5 nm appeared, that did not correspond to heme reoxidation. The absorbance of the α and β bands (572.5 and 540 nm, respectively) also decreased (Fig 3B).
No clear isosbestic points could be detected, indicating complexity. In addition, the patterns of Soret absorbance decrease were different from the pattern of enzyme inactivation (Fig. 1 and Fig. 3 insets), implicating other processes besides heme alteration in inactivation of CBS by peroxynitrite.
Taken together, the spectral changes upon peroxynitrite treatment, particularly the decrease in the 449 nm absorbance of Fe(II)CBS and the appearance of a 424.5 nm peak, suggest loss of thiolate coordination and substitution by a neutral ligand according to previously reported spectra for Cys52 or Arg266 mutants and for native CBS exposed mercuric chloride or reduction/heat [12, 18–20, 22]. The loss of thiolate coordination seems to be accompanied, especially at high peroxynitrite concentrations, by heme degradation involving cleavage of the porphyrin macrocycle, as observed by the bleaching in the Soret and α and β bands.
In the low energy region of the electronic spectrum (Fig. 3C), the changes observed in the ligand-to-metal charge-transfer bands (LMCT band), at 650 and 703 nm, also suggest loss of Cys52 coordination [14]. In addition, a decrease in the αβ bands of Fe(III)CBS at 550 nm was observed. Above 1 mM peroxynitrite, the decreases at 550, 650 and 703 nm were accompanied by an increase at 630 nm, indicating formation of new species.
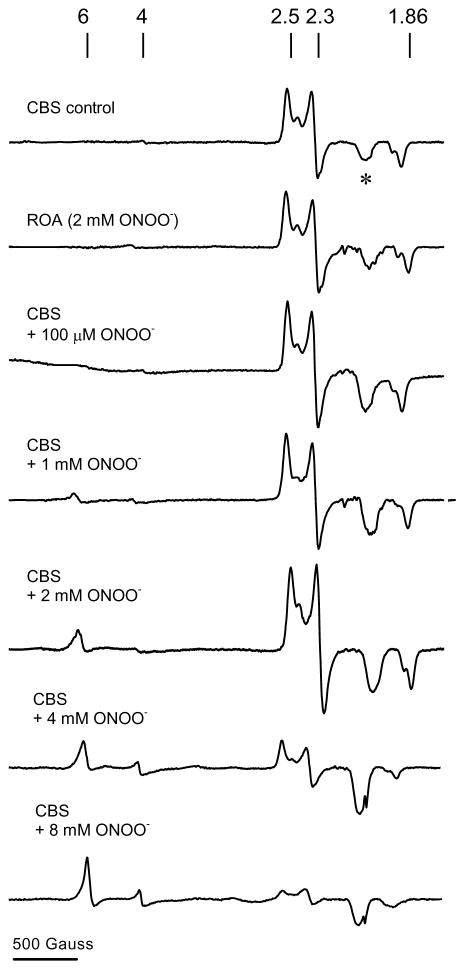
EPR spectrum of ferric CBS exposed to peroxynitrite
The EPR spectrum of Fe(III)CBS exhibits a rhombic signal with g values of 2.5, 2.3 and 1.86, similar to the reported values and characteristic of low-spin heme [12]. Treatment with peroxynitrite resulted in the concentration-dependent appearance of an EPR signal in the g = 6 region, consistent with the conversion of low-spin heme to a high-spin species (Fig. 4). In contrast, no changes in this region were detected upon incubation of CBS with previously decomposed peroxynitrite. The predominance of the high-spin species in the EPR spectrum has been reported for CBS in the presence of mercuric chloride and in Cys52 mutants [12, 20]. This change is consistent with perturbation of the heme ligand environment by peroxynitrite and loss of cysteine coordination, as also indicated by UV-visible spectrophotometry (Fig. 3).
Fig. 4.
EPR spectra of peroxynitrite-treated CBS. Fe(III)CBS (120 μM) was mixed with increasing concentrations of peroxynitrite (0–8 mM) in phosphate buffer (0.1 M, pH 7.4). After 5 min at 37 °C, samples were frozen in liquid nitrogen. The EPR spectra were recorded at 10 K, 1 mW microwave power, 2 × 104 receiver gain, 12.78 G modulation amplitude, 9.393 GHz microwave frequency, and 100 kHz modulation frequency using 1024 data points. Each spectrum represents a single scan. The asterisk denotes a signal present in the cavity. The line markers are at g = 6, 4, 2.5, 2.3, and 1.86, respectively. ROA, reverse order of addition control.
An oxo-ferryl species is not detected
Some heme proteins are able to increase the rate and yield of one-electron processes from peroxynitrite, augmenting nitration through the formation of oxo-ferryl compounds. With Fe(III)CBS (10 μM), an increase in nitration of free tyrosine (0.5 mM) by equimolar peroxynitrite was not detected. Rather, nitration of free tyrosine was inhibited by ~35%, probably due to competition by CBS for nitrating agents (data not shown).
Formation of a transient oxo-ferryl species was probed by stopped-flow spectroscopy in which Fe(III)CBS (5 μM) was mixed with peroxynitrite (130 μM). However, the time-dependent spectra did not show the formation of any detectable oxo-ferryl intermediate concomitant with peroxynitrite decay (Fig. 5). Rather, a decrease in absorbance at 429 nm and 550 nm with an increase at ~380 nm was observed, consistent with the changes previously observed in the end point spectra (Fig. 3).
Fig. 5.
Time-dependent UV-visible spectra of CBS exposed to peroxynitrite. Fe(III)CBS (5 μM) was mixed with peroxynitrite (130 μM) in a stopped-flow spectrophotometer in phosphate buffer (0.1 M, pH 7.4, DTPA 0.1 mM) at 37.0 ± 0.1 °C. Spectra were recorded from 0.0276 to 8.86 s, every 0.0589 s, with an integration time of 3.680 ms.
HPLC and mass spectrometric analysis of the heme
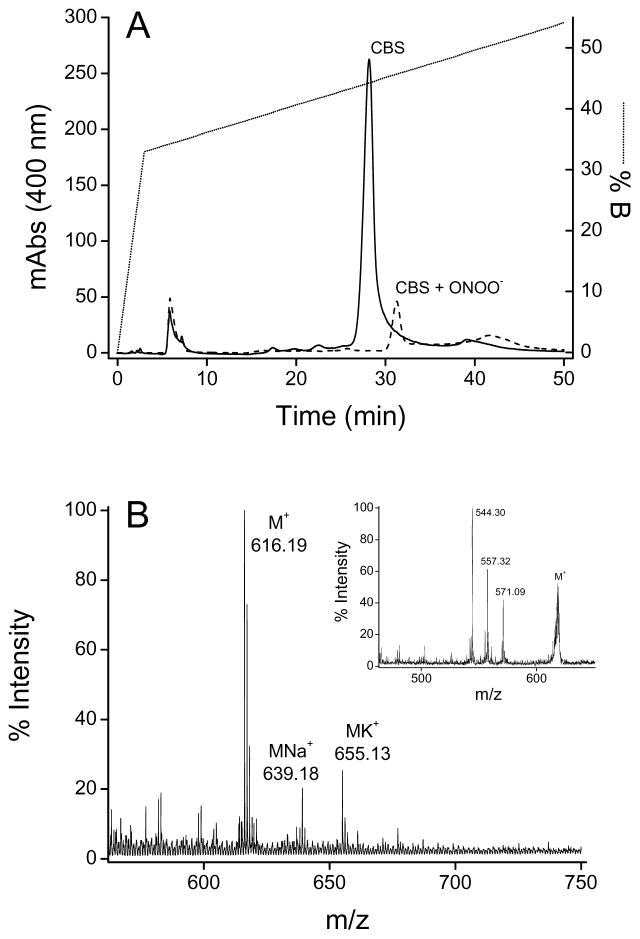
Under the acidic conditions used, the heme in untreated (control) Fe(III)CBS separated from the protein, and was detected by the appearance of a peak with a retention time of 28 min with absorption at 400 nm (Fig. 6A). The MS analysis showed an heme characteristic signal at m/z = 616.19 (theoretical monoisotopic m/z = 616.18 Da) and its distinctive isotopic pattern. This was confirmed through MS/MS fragmentation, by the presence of typical ions generated by losses of formyl (•COOH), acetyl (•CH2COOH) or propionyl (•CH2CH2COOH) fragments from the heme (Fig. 6B) [53].
Fig. 6.
HPLC and mass spectrometric analyses of heme. (A) Fe(III)CBS (5 μM) was treated with peroxynitrite (500 μM). Samples were concentrated, submitted to reverse phase HPLC and analyzed by mass spectrometry. The solid line represents heme isolated from the control CBS sample and the dashed line represents the peroxynitrite-treated sample. The dotted line corresponds to the HPLC gradient (see Materials and methods). (B) MALDI-TOF MS analysis of HPLC-purified heme from control CBS. (Inset) MS/MS spectrum of the parent ion with m/z 616.19 showing the main fragment ions.
The HPLC analysis of peroxynitrite-treated CBS yielded a peak with decreased intensity and a retention time of 31 min (Fig. 6A). The decreased intensity is consistent with the bleaching observed in the UV-visible spectra. No signal could be detected by MS analysis, suggesting the formation of various products. In this regard, products eluting later than native heme upon peroxynitrite exposure have been reported before for cytochrome P450 2B6, where nitroheme was detected [54]. Covalent binding of the heme to the protein could also contribute to explain our results.
Nitration of amino acidic residues
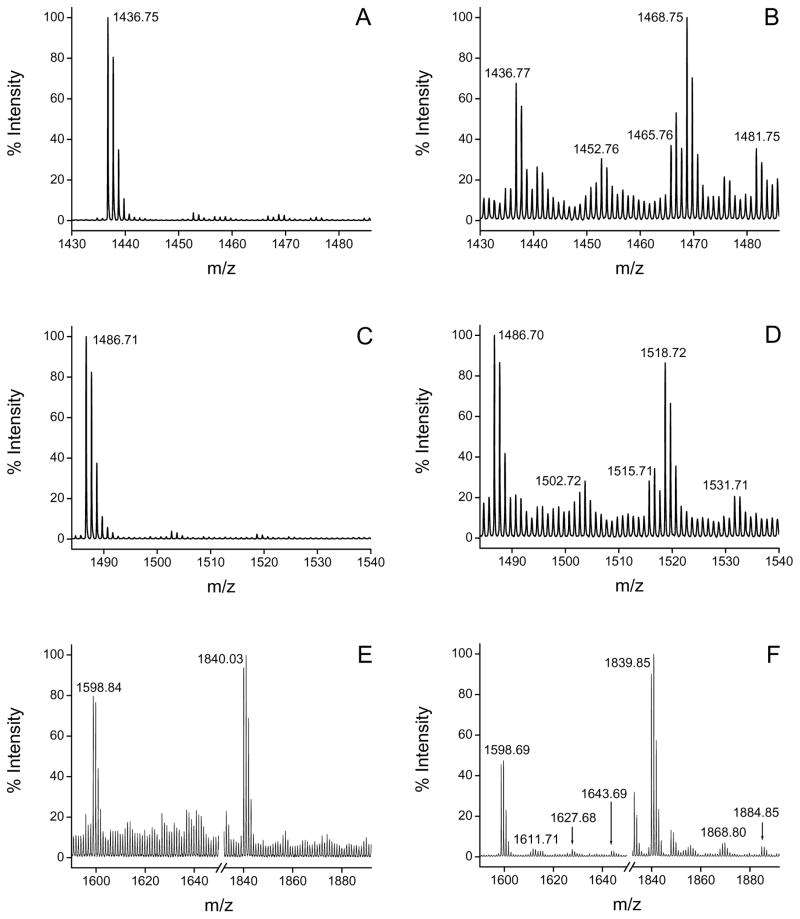
Control and peroxynitrite-treated CBS were digested with trypsin after carbamidomethylation of cysteine residues with iodoacetamide. Four peptides were found to be modified by nitration in the treated sample (Fig. 7 and Table 2), and each of them contained potentially nitratable residues. The peptides with m/z 1436.75 and 1486.71 had in the sequence Trp43 and Trp208, respectively. For the +45 modified peptides, typical nitropeptide photodecomposition was observed, with an ion corresponding to a mass loss of −16 with respect to the +45 ion. Indeed, nitrotyrosine peptides have associated peaks corresponding to +13, +15, +29, +31 and +45 [55]. Peptides with mass increases of +16 and +32 could also be observed in the MS spectra, which could correspond to photodecomposition products of the nitrated peptide or incorporation of one or two oxygens to the native peptide. MS/MS analysis and database search confirmed the sequence of the peptides and the sites of nitration. The fragmentation of the ion of m/z 1481.75 led to the identification of the sequence EPLWIRPDAPSR from human CBS containing a nitrotryptophan residue, with significant ion scores (p < 0.05). In the same way, the MS/MS spectrum of the parent ion of m/z 1531.71 confirmed the presence of nitrotryptophan within the sequence FDSPESHVGVAWR (ion score = 26, p < 0.05). The fragment ions detected for each peptide are shown in Table 3, and allow us to unequivocally identify Trp43 and Trp208 as nitrated residues in the human CBS sequence.
Fig. 7.
Mass spectrometric analysis of tryptic digests of control and peroxynitrite-treated CBS. Fe(III)CBS (5 μM) was exposed to peroxynitrite (500 μM). Samples were carbamidomethylated, trypsinized and subjected to MALDI-TOF MS analysis. Tryptic fragments where modifications were detected are shown for control (left panel, A, C, E) and peroxynitrite-treated CBS (right panel, B, D, F). Modifications were detected for the fragments with m/z 1436.77 (A and B), 1486.70 (C and D) and 1598.69/1839.85 (E and F).
Table 2.
Monoisotopic molecular mass/charge ratios of nitrated peptides and identification of nitration sites
| Peptide From-To | Theoretical am/z | Observed m/z | Assigned sequence | Nitrated residue |
|---|---|---|---|---|
| 40–51 | Native 1436.76 | 1436.77 | EPLWIRPDAPSR b | W43 b |
| 1452.76 | ||||
| 1465.76 | ||||
| 1468.75 | ||||
| Nitrated 1481.75 | 1481.75 | EPLWNO2IRPDAPSR b | ||
| 197–209 | Native 1486.71 | 1486.70 | FDSPESHVGVAWR b | W208 b |
| 1502.72 | ||||
| 1515.71 | ||||
| 1518.72 | ||||
| Nitrated 1531.70 | 1531.71 | FDSPESHVGVAWNO2R b | ||
| 212–224 | Native 1598.79 | 1598.69 | NEIPNSHILDQYR | Y223 |
| 1611.71 | ||||
| 1627.68 | ||||
| Nitrated 1643.78 | 1643.69 | NEIPNSHILDQYNO2R | ||
| 210–224 | Native 1839.97 | 1839.85 | LKNEIPNSHILDQYR | Y223 |
| 1868.80 | ||||
| Nitrated 1884.96 | 1884.85 | LKNEIPNSHILDQYNO2R | ||
The nitro group results in a mass increase of 44.99 Da.
The sequence and nitration site were confirmed by MS/MS analysis.
Table 3.
Results of protein database search with MS/MS spectrum data
| (A) Sequence matched to MS/MS data generated from precursor ion m/z 1481.75. Mascot ion score = 19 (p < 0.05). | ||||
|---|---|---|---|---|
| b | AA | y | ||
| 130.05 | 1 | E | 12 | |
| 227.1 | 2 | P | 11 | 1352.71 |
| 340.19 | 3 | L | 10 | 1255.65 |
| 571.25 | 4 | WNO2 | 9 | 1142.57 |
| 684.34 | 5 | I | 8 | 911.51 |
| 840.44 | 6 | R | 7 | 798.42 |
| 937.49 | 7 | P | 6 | 642.32 |
| 1052.52 | 8 | D | 5 | 545.27 |
| 1123.55 | 9 | A | 4 | 430.24 |
| 1220.61 | 10 | P | 3 | 359.2 |
| 1307.64 | 11 | S | 2 | 262.15 |
| 12 | R | 1 | 175.12 | |
| (B) Sequence matched to MS/MS data generated from precursor ion m/z 1531.71. Mascot ion score = 26 (p < 0.05). | ||||
|---|---|---|---|---|
| b | AA | y | ||
| 148.08 | 1 | F | 13 | |
| 263.1 | 2 | D | 12 | 1384.62 |
| 350.13 | 3 | S | 11 | 1269.6 |
| 447.19 | 4 | P | 10 | 1182.57 |
| 576.23 | 5 | E | 9 | 1085.51 |
| 663.26 | 6 | S | 8 | 956.47 |
| 800.32 | 7 | H | 7 | 869.44 |
| 899.39 | 8 | V | 6 | 732.38 |
| 956.41 | 9 | G | 5 | 633.31 |
| 1055.48 | 10 | V | 4 | 576.29 |
| 1126.52 | 11 | A | 3 | 477.22 |
| 1357.58 | 12 | WNO2 | 2 | 406.18 |
| 13 | R | 1 | 175.12 | |
N-terminal b and C-terminal y ions are listed. Ions detected in the MS/MS spectrum are shown in bold. AA, amino acids.
The third peptide modified in the peroxynitrite-treated sample (m/z 1598.84) contains Tyr223. In this case, peptides with mass increases of +45, +29 and +13 could be observed. A similar signature pattern was observed for the peptide with m/z 1840.03, which also harbored Tyr223 and resulted from incomplete trypsin cleavage at an internal lysine providing further evidence for its nitration.
In addition to the mass spectrometry data, nitration of tryptophan and tyrosine residues was also suggested by losses in tryptophan fluorescence upon peroxynitrite treatment and by Western blot analysis using a polyclonal anti-nitrotyrosine antibody (data not shown).
Finally, a fragment with m/z 2368.3 could be observed only in the peroxynitrite-treated sample (data not shown). This fragment had the expected mass of the carbamidomethylated tryptic peptide spanning residues 52–72 and encompassing the heme ligand residues, Cys52 and His65. The lack of observation of this peptide in the control sample suggests that Cys52 could not be modified by iodoacetamide when bound to the heme, and can be interpreted as further evidence for loss of Cys52 coordination to the heme with peroxynitrite treatment, in addition to UV-visible and EPR spectral evidence.
Discussion
In this study, we have characterized the inactivation of CBS by peroxynitrite. Based on the kinetic and scavenging data, the loss of CBS activity results from its reaction with peroxynitrite itself and with its derived radicals, nitrogen dioxide and carbonate radical, generated in the presence of carbon dioxide. The global direct second-order rate constant of Fe(III)CBS with peroxynitrite, (2.4–5.0) × 104 M−1 s−1 (pH 7.4, 37 °C), is comparable to those of other heme proteins like Fe(II)cyt c [49], oxyhemoglobin (1.7 × 104 M−1 s−1) [56] and methemoglobin (3.9 × 104 M−1 s−1) [31], higher than Fe(III)cyt c and neuroglobin (undetected direct reaction) [57 M, 58], but lower than hemeperoxidases (106–107 M−1 s−1) [28, 59], nitric oxide synthase (2.2 × 105 M−1 s−1) [32] and Cyt P450BM3 (1 × 106 M−1 s−1) [29]. Based on this second-order rate constant and on the relatively high IC50 of ~150 μM for 5 μM CBS, this enzyme is probably not a preferential target for peroxynitrite in vivo, unless generated at high local concentrations.
The biochemical basis for the inactivation appears to involve several factors. Although PLP, in principle, could be a target for peroxynitrite due to the reactivity of aldehydes, and hence aldimines [60] and the pyridine ring, PLP modification in CBS was not observed. Instead, alterations at the heme level and nitration of amino acidic residues are associated with the inactivation.
The alterations at the heme level involved loss of thiolate coordination, conversion from low spin to high spin and bleaching, without detectable formation of high valent oxo-ferryl compounds or promotion of one-electron chemistry of peroxynitrite. No particular heme modification could be detected through our HPLC/MS analysis, suggesting the formation of a mixture of products. Candidate degradation intermediates could be hydroxyheme, verdoheme, nitroheme and covalent linkages to the protein. In control CBS samples, the acidic treatment followed by reverse phase HPLC provided, for the first time, a procedure to release the very stable ferric heme from the native CBS protein.
The reaction of the heme of Fe(III)CBS with peroxynitrite is reminiscent of that of ferric cytochrome c, which is six-coordinate with Met/His ligands. In this case, bleaching of the Soret band is observed, but not an oxo-ferryl species [57]. With other hydroperoxides, loss of Met80 coordination and conversion from low spin to high spin heme has been reported [61]. The reaction of Fe(III)CBS with peroxynitrite is also reminiscent of the reaction of ferric neuroglobin, another six-coordinated protein with bis-His ligation where formation of a high valent oxo-ferryl species is not observed either [58]. In contrast to these six-coordinated proteins, five-coordinated ferric hemeproteins such as peroxidases, cytochrome P450 and nitric oxide synthase are able to react directly with peroxynitrite, forming oxo-ferryl compounds [28–30, 32]. Thus, our results highlight the importance of coordination state in determining ferric heme reactivity towards peroxynitrite.
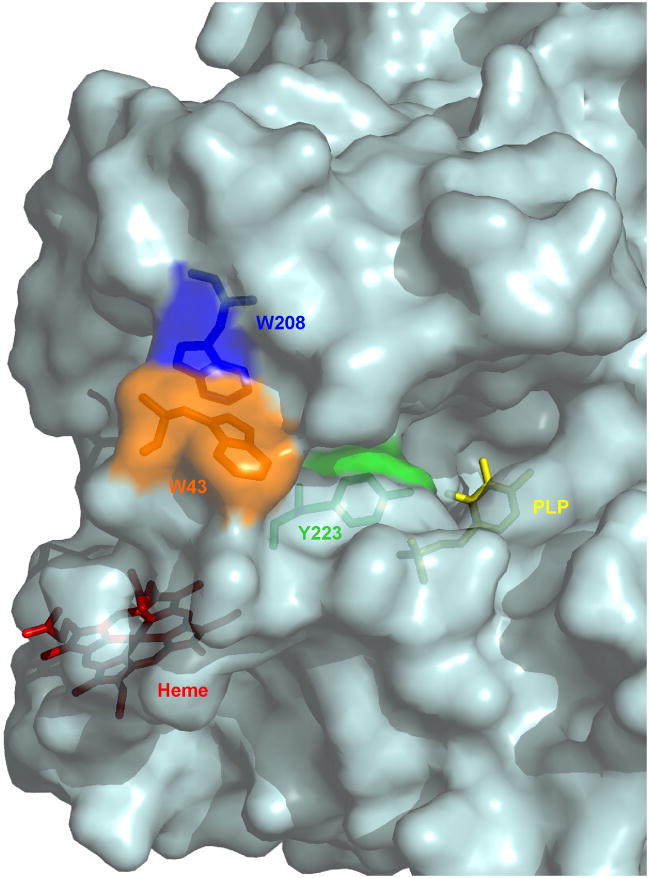
In addition to alterations at the heme level, nitration of protein residues also appear to contibute to inactivation of CBS. Nitration of Trp208, Trp43 and Tyr223 was detected by mass spectrometry. Trp208 is superficial and close to the substrate-binding site. The available structures suggest that Trp43, in the N-terminal end, is superficial and close to Trp208 (Fig. 8 [10, 11]). Tyr223 is buried and has been proposed to be a key residue for substrate specificity, being spatially adjacent to the homocysteine-binding site (Fig. 8 [10, 11]). Introduction of a bulky nitro group at this position, which can lower the pKA of the phenolic group, could interfere with enzyme activity. Furthermore, Tyr223 is adjacent to Arg224, a residue that interacts with the heme carboxylates and causes hereditary hyperhomocysteinemia when mutated.
Fig. 8.
Three-dimensional structure of CBS showing the location of nitrated residues, heme and PLP. The protein is shown in surface representation, with transparency. In stick display, Trp43 is shown in orange, Trp208 in blue, Tyr223 in green, PLP in yellow and the heme in red. The structure was generated using the PDB file 1JBQ [10] and PyMol v0.99 [64].
In summary, our study provides biochemical insights into the properties of the very stable ferric heme in human CBS, which has previously only shown to be sensitive to mercuric chloride. In addition, these results contribute to the general understanding of the interactions of peroxynitrite with hemeproteins. Last, our study provides a mechanistic explanation for the reported decrease in CBS activity and increase in homocysteine that is observed in vivo in rat kidney ischemia-reperfusion injury, which correlates with reactive oxygen/nitrogen species formation and nitration [62, 63]. The inactivation of human CBS by peroxynitrite thus highlights its potential sensitivity to oxidative stress with possible relevance to hyperhomocysteinemia.
Acknowledgments
This work was supported by grants from CSIC (Universidad de la República, Uruguay), Programa de Desarrollo Tecnológico II (Uruguayan Research Council), Programa para el Desarrollo de las Ciencias Básicas (PEDECIBA, Uruguay) and the National Institutes of Health (HL58984). We thank Gerardo Ferrer-Sueta and Rafael Radi for helpful insights, Ernesto Cuevasanta and Matías Möller for technical assistance (Universidad de la República, Montevideo) and Javier Seravelli for help with EPR spectroscopy (University of Nebraska, Lincoln).
Footnotes
Abbreviations used: CBS, cystathionine β-synthase; SAM, S-adenosyl-L-methionine; PLP, pyridoxal 5′-phosphate; DTPA, diethylenetriaminepentaacetic acid; ROA, reverse order of addition; DTNB, 5,5′-dithiobis-(2-nitrobenzoic acid); LMCT, ligand to metal charge transfer band; EPR, electron paramagnetic resonance or electron spin resonance; p-HPA, p-hydroxyphenylacetic acid; HPLC, high performance liquid chromatography; MALDI, matrix-assisted laser desorption ionization; TOF, time-of-flight; MS, mass spectrometry; IC50, concentration of peroxynitrite needed to inactivate by 50%.
The term peroxynitrite is used to refer to both peroxynitrite anion (ONOO−) and peroxynitrous acid (ONOOH). IUPAC-recommended names are oxoperoxonitrate(1-) and hydrogen oxoperoxonitrate, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abe K, Kimura H. J Neurosci. 1996;16:1066–1071. doi: 10.1523/JNEUROSCI.16-03-01066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCully KS. Am J Pathol. 1969;56:111–128. [PMC free article] [PubMed] [Google Scholar]
- 3.Mills JL, McPartlin JM, Kirke PN, Lee YJ, Conley MR, Weir DG, Scott JM. Lancet. 1995;345:149–151. doi: 10.1016/s0140-6736(95)90165-5. [DOI] [PubMed] [Google Scholar]
- 4.Refsum H, Ueland PM, Nygard O, Vollset SE. Annu Rev Med. 1998;49:31–62. doi: 10.1146/annurev.med.49.1.31. [DOI] [PubMed] [Google Scholar]
- 5.Kery V, Poneleit L, Kraus JP. Arch Biochem Biophys. 1998;355:222–232. doi: 10.1006/abbi.1998.0723. [DOI] [PubMed] [Google Scholar]
- 6.Taoka S, Widjaja L, Banerjee R. Biochemistry. 1999;38:13155–13161. doi: 10.1021/bi990865t. [DOI] [PubMed] [Google Scholar]
- 7.Finkelstein JD, Kyle WE, Martin JL, Pick AM. Biochem Biophys Res Commun. 1975;66:81–87. doi: 10.1016/s0006-291x(75)80297-x. [DOI] [PubMed] [Google Scholar]
- 8.Zou CG, Banerjee R. J Biol Chem. 2003;278:16802–16808. doi: 10.1074/jbc.M212376200. [DOI] [PubMed] [Google Scholar]
- 9.Kery V, Bukovska G, Kraus JP. J Biol Chem. 1994;269:25283–25288. [PubMed] [Google Scholar]
- 10.Meier M, Janosik M, Kery V, Kraus JP, Burkhard P. Embo J. 2001;20:3910–3916. doi: 10.1093/emboj/20.15.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taoka S, Lepore BW, Kabil O, Ojha S, Ringe D, Banerjee R. Biochemistry. 2002;41:10454–10461. doi: 10.1021/bi026052d. [DOI] [PubMed] [Google Scholar]
- 12.Ojha S, Hwang J, Kabil O, Penner-Hahn JE, Banerjee R. Biochemistry. 2000;39:10542–10547. doi: 10.1021/bi000831h. [DOI] [PubMed] [Google Scholar]
- 13.Taoka S, Ohja S, Shan X, Kruger WD, Banerjee R. J Biol Chem. 1998;273:25179–25184. doi: 10.1074/jbc.273.39.25179. [DOI] [PubMed] [Google Scholar]
- 14.Pazicni S, Lukat-Rodgers GS, Oliveriusova J, Rees KA, Parks RB, Clark RW, Rodgers KR, Kraus JP, Burstyn JN. Biochemistry. 2004;43:14684–14695. doi: 10.1021/bi0488496. [DOI] [PubMed] [Google Scholar]
- 15.Carballal S, Madzelan P, Zinola CF, Grana M, Radi R, Banerjee R, Alvarez B. Biochemistry. 2008;47:3194–3201. doi: 10.1021/bi700912k. [DOI] [PubMed] [Google Scholar]
- 16.Taoka S, West M, Banerjee R. Biochemistry. 1999;38:2738–2744. doi: 10.1021/bi9826052. [DOI] [PubMed] [Google Scholar]
- 17.Vadon-Le Goff S, Delaforge M, Boucher JL, Janosik M, Kraus JP, Mansuy D. Biochem Biophys Res Commun. 2001;283:487–492. doi: 10.1006/bbrc.2001.4807. [DOI] [PubMed] [Google Scholar]
- 18.Taoka S, Green EL, Loehr TM, Banerjee R. J Inorg Biochem. 2001;87:253–259. doi: 10.1016/s0162-0134(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 19.Pazicni S, Cherney MM, Lukat-Rodgers GS, Oliveriusova J, Rodgers KR, Kraus JP, Burstyn JN. Biochemistry. 2005;44:16785–16795. doi: 10.1021/bi051305z. [DOI] [PubMed] [Google Scholar]
- 20.Ojha S, Wu J, LoBrutto R, Banerjee R. Biochemistry. 2002;41:4649–4654. doi: 10.1021/bi011827o. [DOI] [PubMed] [Google Scholar]
- 21.Evande R, Ojha S, Banerjee R. Arch Biochem Biophys. 2004;427:188–196. doi: 10.1016/j.abb.2004.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Singh S, Madzelan P, Stasser J, Weeks CL, Becker D, Spiro TG, Penner-Hahn J, Banerjee R. J Inorg Biochem. 2009;103:689–697. doi: 10.1016/j.jinorgbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Radi R, Beckman JS, Bush KM, Freeman BA. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 24.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Proc Natl Acad Sci U S A. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer-Sueta G, Radi R. ACS Chem Biol. 2009;4:161–177. doi: 10.1021/cb800279q. [DOI] [PubMed] [Google Scholar]
- 26.Trujillo M, Alvarez B, Souza JM, Romero N, Castro L, Thomson L, Radi R. In: Nitric Oxide: Biology and Pathobiology. 2. Ignarro JL, editor. Elsevier; 2009. in press. [Google Scholar]
- 27.Denicola A, Freeman BA, Trujillo M, Radi R. Arch Biochem Biophys. 1996;333:49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- 28.Floris R, Piersma SR, Yang G, Jones P, Wever R. Eur J Biochem. 1993;215:767–775. doi: 10.1111/j.1432-1033.1993.tb18091.x. [DOI] [PubMed] [Google Scholar]
- 29.Daiber A, Herold S, Schoneich C, Namgaladze D, Peterson JA, Ullrich V. Eur J Biochem. 2000;267:6729–6739. doi: 10.1046/j.1432-1033.2000.01768.x. [DOI] [PubMed] [Google Scholar]
- 30.Sheng X, Horner JH, Newcomb M. J Am Chem Soc. 2008;130:13310–13320. doi: 10.1021/ja802652b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold S, Shivashankar K. Biochemistry. 2003;42:14036–14046. doi: 10.1021/bi0350349. [DOI] [PubMed] [Google Scholar]
- 32.Marechal A, Mattioli TA, Stuehr DJ, Santolini J. J Biol Chem. 2007;282:14101–14112. doi: 10.1074/jbc.M609237200. [DOI] [PubMed] [Google Scholar]
- 33.Radi R, Peluffo G, Alvarez MN, Naviliat M, Cayota A. Free Radic Biol Med. 2001;30:463–488. doi: 10.1016/s0891-5849(00)00373-7. [DOI] [PubMed] [Google Scholar]
- 34.Peluffo G, Radi R. Cardiovasc Res. 2007;75:291–302. doi: 10.1016/j.cardiores.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 35.Saha A, Goldstein S, Cabelli D, Czapski G. Free Radic Biol Med. 1998;24:653–659. doi: 10.1016/s0891-5849(97)00365-1. [DOI] [PubMed] [Google Scholar]
- 36.Hughes MN, Nicklin HG. J Chem Soc A. 1968:450–452. [Google Scholar]
- 37.Green LC, Wagner DA, Glogowski J, Skipper PL, Wishnok JS, Tannenbaum SR. Anal Biochem. 1982;126:131–138. doi: 10.1016/0003-2697(82)90118-x. [DOI] [PubMed] [Google Scholar]
- 38.Shan X, Kruger WD. Nat Genet. 1998;19:91–93. doi: 10.1038/ng0598-91. [DOI] [PubMed] [Google Scholar]
- 39.Kashiwamata S, Greenberg DM. Biochim Biophys Acta. 1970;212:488–500. doi: 10.1016/0005-2744(70)90255-x. [DOI] [PubMed] [Google Scholar]
- 40.Ellman G, Lysko H. Anal Biochem. 1979;93:98–102. [PubMed] [Google Scholar]
- 41.Schellenberg KAHL. J Biol Chem. 1958;231:547–556. [PubMed] [Google Scholar]
- 42.Alvarez B, Ferrer-Sueta G, Freeman BA, Radi R, Alvarez B, Ferrer-Sueta G, Freeman BA, Radi R. J Biol Chem. 1999;274:842–848. doi: 10.1074/jbc.274.2.842. [DOI] [PubMed] [Google Scholar]
- 43.Quijano C, Hernandez-Saavedra D, Castro L, McCord JM, Freeman BA, Radi R. J Biol Chem. 2001;276:11631–11638. doi: 10.1074/jbc.M009429200. [DOI] [PubMed] [Google Scholar]
- 44.Massey V. Biochim Biophys Acta. 1959;34:255–256. doi: 10.1016/0006-3002(59)90259-8. [DOI] [PubMed] [Google Scholar]
- 45.Beckman JS, Ischiropoulos H, Zhu L, van der Woerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Arch Biochem Biophys. 1992;298:438–445. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 46.Colas C, Kuo JM, Ortiz de Montellano PR. J Biol Chem. 2002;277:7191–7200. doi: 10.1074/jbc.M109523200. [DOI] [PubMed] [Google Scholar]
- 47.Alvarez B, Demicheli V, Duran R, Trujillo M, Cervenansky C, Freeman BA, Radi R. Free Radic Biol Med. 2004;37:813–822. doi: 10.1016/j.freeradbiomed.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Blanchard-Fillion B, Souza JM, Friel T, Jiang GC, Vrana K, Sharov V, Barron L, Schoneich C, Quijano C, Alvarez B, Radi R, Przedborski S, Fernando GS, Horwitz J, Ischiropoulos H. J Biol Chem. 2001;276:46017–46023. doi: 10.1074/jbc.M105564200. [DOI] [PubMed] [Google Scholar]
- 49.Thomson L, Trujillo M, Telleri R, Radi R. Arch Biochem Biophys. 1995;319:491–497. doi: 10.1006/abbi.1995.1321. [DOI] [PubMed] [Google Scholar]
- 50.Espenson JH. Chemical kinetics and reaction mechanisms. 2. McGraw-Hill; 1995. [Google Scholar]
- 51.Kery V, Poneleit L, Meyer JD, Manning MC, Kraus JP. Biochemistry. 1999;38:2716–2724. doi: 10.1021/bi981808n. [DOI] [PubMed] [Google Scholar]
- 52.Frank N, Kery V, Maclean KN, Kraus JP. Biochemistry. 2006;45:11021–11029. doi: 10.1021/bi060737m. [DOI] [PubMed] [Google Scholar]
- 53.Demirev PA, Feldman AB, Kongkasuriyachai D, Scholl P, Sullivan D, Jr, Kumar N. Anal Chem. 2002;74:3262–3266. doi: 10.1021/ac025621k. [DOI] [PubMed] [Google Scholar]
- 54.Lin HL, Myshkin E, Waskell L, Hollenberg PF. Chem Res Toxicol. 2007;20:1612–1622. doi: 10.1021/tx700220e. [DOI] [PubMed] [Google Scholar]
- 55.Sarver A, Scheffler NK, Shetlar MD, Gibson BW. J Am Soc Mass Spectrom. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]
- 56.Denicola A, Souza JM, Radi R. Proc Natl Acad Sci U S A. 1998;95:3566–3571. doi: 10.1073/pnas.95.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gebicka L, Didik J. Acta Biochim Pol. 2003;50:815–823. [PubMed] [Google Scholar]
- 58.Herold S, Fago A, Weber RE, Dewilde S, Moens L. J Biol Chem. 2004;279:22841–22847. doi: 10.1074/jbc.M313732200. [DOI] [PubMed] [Google Scholar]
- 59.Furtmuller PG, Jantschko W, Zederbauer M, Schwanninger M, Jakopitsch C, Herold S, Koppenol WH, Obinger C. Biochem Biophys Res Commun. 2005;337:944–954. doi: 10.1016/j.bbrc.2005.09.138. [DOI] [PubMed] [Google Scholar]
- 60.Uppu RM, Winston GW, Pryor WA. Chem Res Toxicol. 1997;10:1331–1337. doi: 10.1021/tx970056f. [DOI] [PubMed] [Google Scholar]
- 61.Nantes IL, Faljoni-Alario A, Nascimento OR, Bandy B, Gatti R, Bechara EJ. Free Radic Biol Med. 2000;28:786–796. doi: 10.1016/s0891-5849(00)00170-2. [DOI] [PubMed] [Google Scholar]
- 62.Prathapasinghe GA, Siow YL, KO Am J Physiol Renal Physiol. 2007;292:F1354–1363. doi: 10.1152/ajprenal.00301.2006. [DOI] [PubMed] [Google Scholar]
- 63.Prathapasinghe GA, Siow YL, Xu Z, KO Am J Physiol Renal Physiol. 2008;295:F912–922. doi: 10.1152/ajprenal.00040.2008. [DOI] [PubMed] [Google Scholar]
- 64.DeLano WL. DeLano Scientific. San Carlos; CA, USA: 2002. http://www.pymol.org. [Google Scholar]