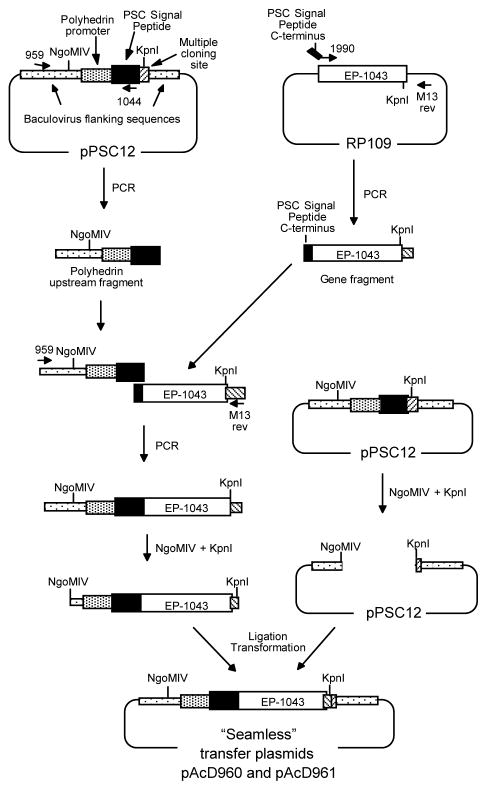
Figure 2. Overlap extension synthesis and cloning strategy for insertion of the EP-1043 HTL epitope coding sequence into the baculovirus transfer plasmid.
The EP-1043 gene product was synthesized using overlapping oligonucleotides, averaging 60 to 90 bp in length with overlaps of approximately 15–20 bp, (Operon Technologies, Alameda, CA). Constructs were assembled by extending the overlapping oligonucleotides using pfu polymerase (Stratagene, San Diego). The resulting full-length product was sequenced and sub-cloned into RP109 DNA plasmid vector. The EP-1043 DNA plasmid was used as a template to fuse a segment of the AcNPV signal peptide to the HTL epitope coding sequence using the O-1990 5′ primer (GGTCGCCGTTTCTAACGCG/GAAAAAGTCTACCTGGCATGG: the C-terminus of the signal peptide/N-terminus of the EP-1043 HTL epitope gene) and the M13rev 3′ primer (TCACACAGGAAACAGCTATGAC: pCR Blunt downstream). The resulting 1.1 kbp PCR product was purified. The pPSC12 plasmid consists of the AcNPV EcoRI “I” fragment inserted into pUC8, with the polyhedrin coding sequences 3′ of the ATG start codon replaced with the AcNPN signal peptide and a polylinker site. The pPSC12 plasmid was used as a PCR template with the O-959 5′ primer (CTGGTAGTTCTTCGGAGTGTG: polyhedron upstream region) which annealed upstream of the unique NgoMIV and EcoRV sites and the O-1044 3′ primer (CGCGTTAGAAACGGCGACC: C-terminus of the signal peptide) which annealed to the 3′ end to produce the PSC signal peptide. To fuse the PSC signal peptide sequence to EP-1043, another PCR reaction was run. Using primers O-959 and M13rev, 1.1kb EP-1043 was combined with PSC signal peptide annealed and extended by PCR. The resulting PCR product was digested with NgoMIV and KpnI, the 1.6 kbp fragment was purified and ligated into NgoMIV-KpnI-cut pPSC12.