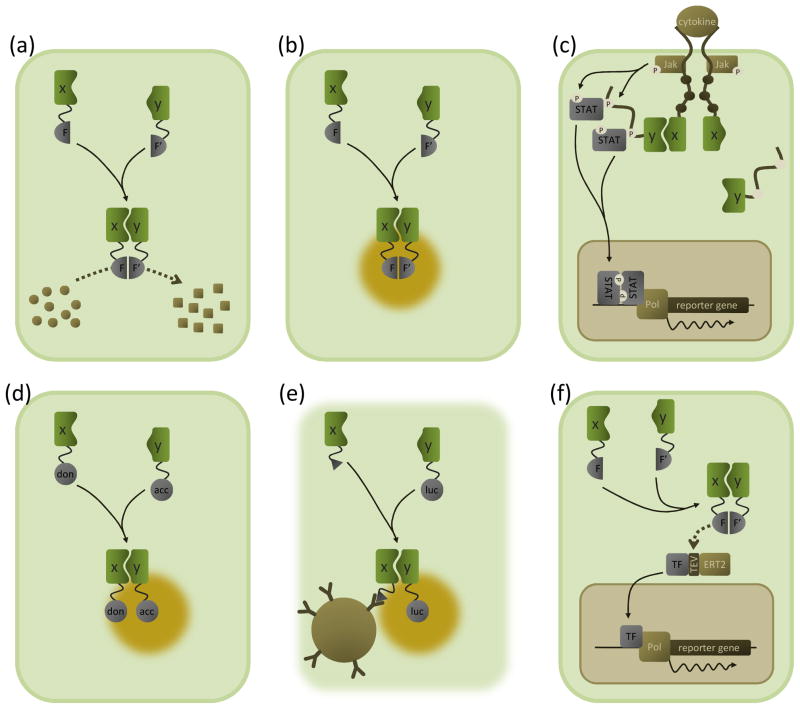
Figure 1. Overview of M2H assay principles.
Schematic outline of the M2H assays discussed in the text. The interacting proteins X and Y are depicted in green; complemented protein fragments or proteins are grey. Please note that the M2H (sensu stricto) assay is not depicted here as it is conceptually identical to the Y2H system (Box 1).
(a) Enzyme complementation systems. The interaction between two proteins of interest (X and Y) brings together two fragments of a reporter enzyme (F and F′); this reconstitutes the enzyme’s catalytic activity, converting a substrate (circles) into a detectable product (squares). The enzyme fragments can be pre-folded subunits (e.g. the split β-galactosidase assay) or fragments that fold only upon complementation (e.g. the β-lactamase and luciferase protein fragment complementation assays).
(b) BiFC. Association of proteins X and Y reconstitutes a fluorescent protein from two fragments (F and F′), resulting in fluorescent emission upon excitation at a suitable wavelength (yellow halo).
(c) MAPPIT. Bait X is coupled to a signalling-deficient cytokine receptor (via substituting tyrosine residues in the receptor tail which are critical for recruiting signal transducer and activator of transcription (STAT) proteins (brown dots)). The prey Y is tethered to a portion of another receptor containing intact recruitment sites. Upon bait–prey interaction, a functional receptor is reconstituted which can be activated by the appropriate cytokine ligand. Upon ligand binding, pre-associated Janus kinases (Jak) are activated by cross-phosphorylation (P). Activated Jaks phosphorylate (P) tyrosine residues in the receptor fragment coupled to the prey (white dots), which then act as docking sites for STATs. Recruited STATs are in turn phosphorylated by the Jaks, leading to their activation and subsequent dissociation and translocation to the nucleus. In the nucleus, STAT dimers induce STAT-dependent reporter gene transcription. In the MASPIT setup, dihydrofolate reductase (DHFR) is coupled to the signalling deficient receptor chain and a chemical compound is fused to methotrexate, a molecule that binds to DHFR with very high affinity. Addition of such a methotrexate fusion compound to cells expressing the DHFR-coupled receptor results in the chemical compound being displayed as a bait.
(d) FRET and BRET. Association of proteins X and Y brings into proximity an energy donor (don) and acceptor protein (acc). In FRET, (intact) fluorescent proteins are used as donor and acceptor. Excitation of the donor fluorophore causes nonradiative energy transfer to occur between the two, resulting in fluorescent emission of the acceptor fluorophore (yellow halo). Instead of a fluorescent protein, a luciferase enzyme is used as a donor in BRET. Enzymatic oxidation of a luciferase substrate generates bioluminescence, and energy transfer to the acceptor protein produces a fluorescent signal.
(e) LUMIER. Interaction between a Flag-tagged (triangle) bait protein X and a prey protein Y fused to Renilla luciferase is detected by bioluminescence measurements (yellow halo) of immunoprecipitates obtained from cell lysates using anti-Flag antibodies coupled to sepharose beads (brown sphere).
(f) Split TEV assay. Association between proteins X and Y complements a functional TEV protease from fragments F and F′. At the site of a TEV recognition sequence, the protease cleaves a chimeric protein consisting of a transcription factor (TF) coupled to an ERT2 domain, thus retaining the transcription factor in the cytosol. After cleavage, the released transcription factor migrates into the nucleus where it induces reporter gene transcription. Note that the split TEV assay is a flexible system and that the setup shown here is only one of the possible choices.