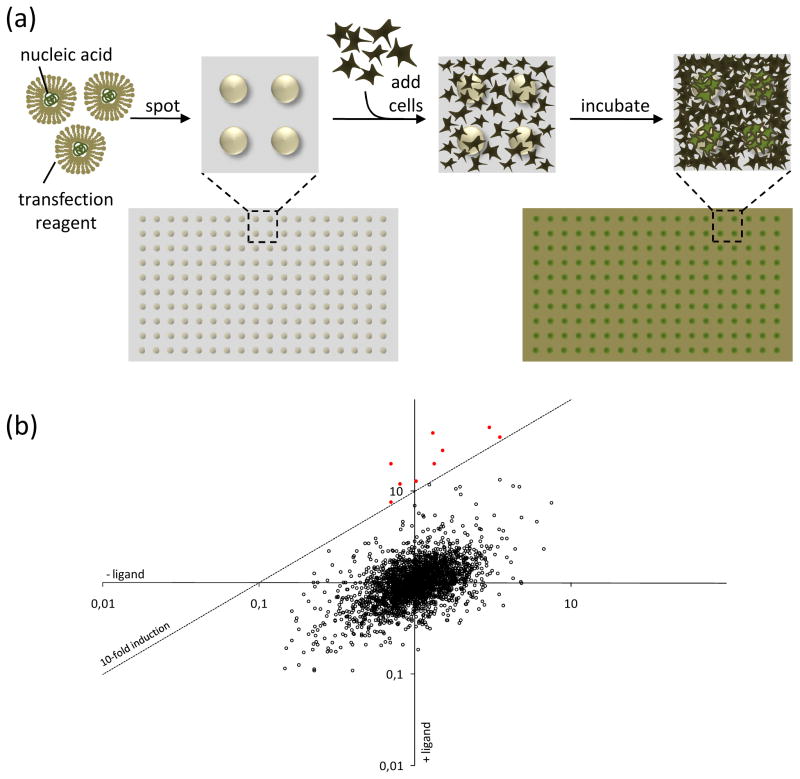
Figure I. Reverse transfected cell arrays and their application in MAPPIT.
(a) Generation of a cell array. Reverse transfection mixtures containing nucleic acids complexed with a transfection reagent are spotted onto a carrier surface. After drying, these arrays are overlaid with cells and incubated. Cells growing on top of the spots are (reverse) transfected with the nucleic acids present in the dried reverse transfection complexes, resulting in an array of transfected cells.
(b) Result of an array MAPPIT screen. MAPPIT is an inducible assay that requires stimulation with an appropriate cytokine ligand. To exploit this additional level of control, luciferase signals obtained from samples treated with ligand are corrected for the background signal from samples that were not ligand-stimulated. The resulting induction factor is used to score interactions. In the conceptual example depicted here, each dot represents a different prey being combined with the bait that is being screened. For each bait–prey combination the normalized luciferase activity obtained with and without addition of the ligand are plotted against each other. Bait–prey combinations yielding a signal above a user-defined cut-off (dashed line; here corresponding to 10-fold induction of ligand treated versus untreated) score positive (red dots).