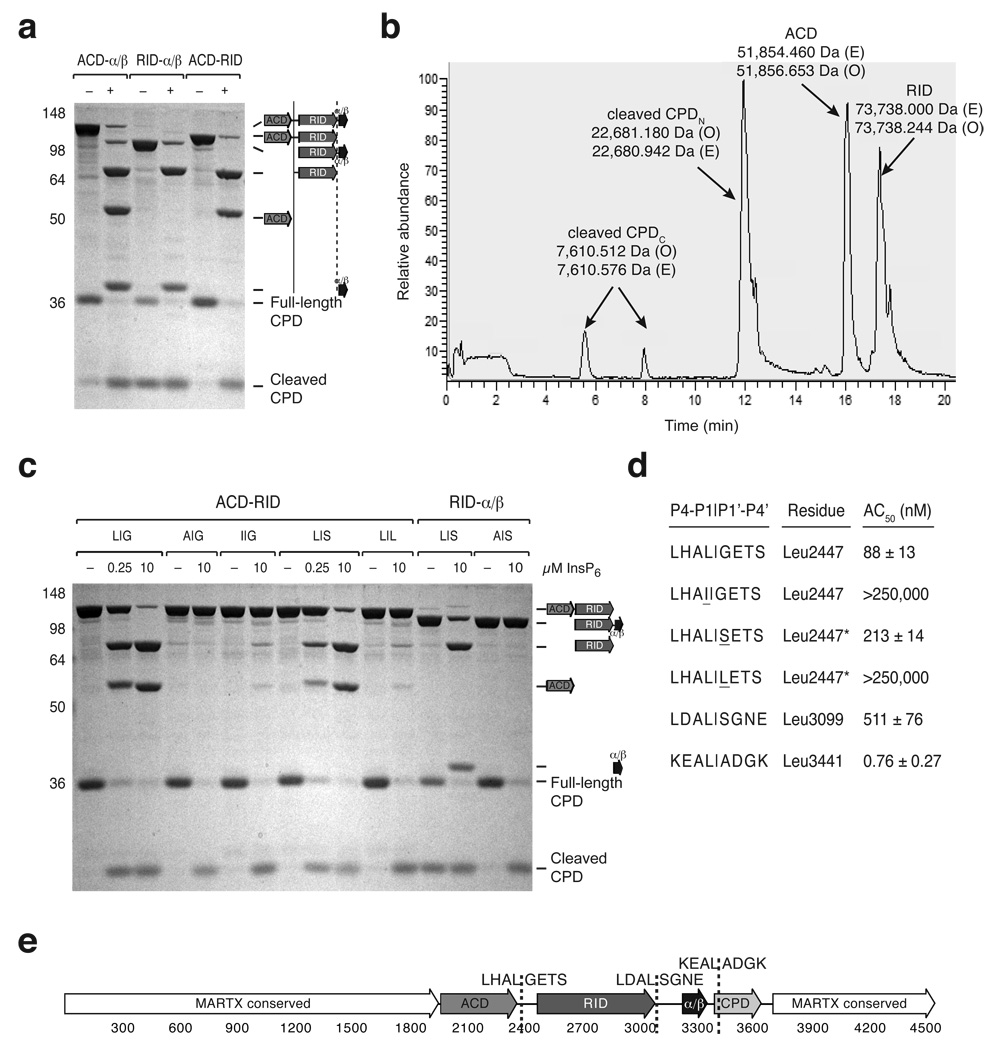
Figure 5.
Identification of MARTXVc toxin cleavage sites in vitro. (a) CPD-mediated transcleavage of MARTXVc polypeptides in vitro. Recombinant CPD (aa 3391–3650) and the indicated MARTXVc polypeptides were incubated ± InsP6, and cleavage reactions were resolved by SDS-PAGE and visualized by SDS-PAGE (schematic shown to the right). (b) Reverse-phase HPLC chromatogram of CPD-mediated transcleavage of recombinant ACD-RID. The observed masses (O) of the polypeptides detected within a given peak are indicated and were determined by FT-MS analysis; the expected masses (E) of polypeptide fragments are also shown. (c) InsP6-induced CPD-mediated transcleavage of mutant MARTXVc polypeptides. The P1 and P1’ residues of the wildtype and mutant cleavage sites are given as P1|P1’. L|G is the wildtype sequence for the ACD-RID cleavage site (Leu2447); L|S is the wildtype sequence for the RID-α/β cleavage site (Leu3099). (d) Comparison of CPD-mediated cleavage at various processing sites. The P4 to P4’ residues are shown for each cleavage site. The concentration of InsP6 at which 50% cleavage of the indicated polypeptides occurred (AC50) is shown (± s.d.). In the presence of InsP6, the ACD-RID and RID-α/β polypeptides were subjected to transcleavage with recombinant CPD, while the α/β-CPD polypeptide was subjected to autocleavage. Mutations introduced into either the P1 and P1’ sites of the ACD-RID cleavage site are underlined. (e) Schematic of MARTXVc toxin. Amino acid numbering is given below, and cleavage sites sequences (P4-P4’) are given.