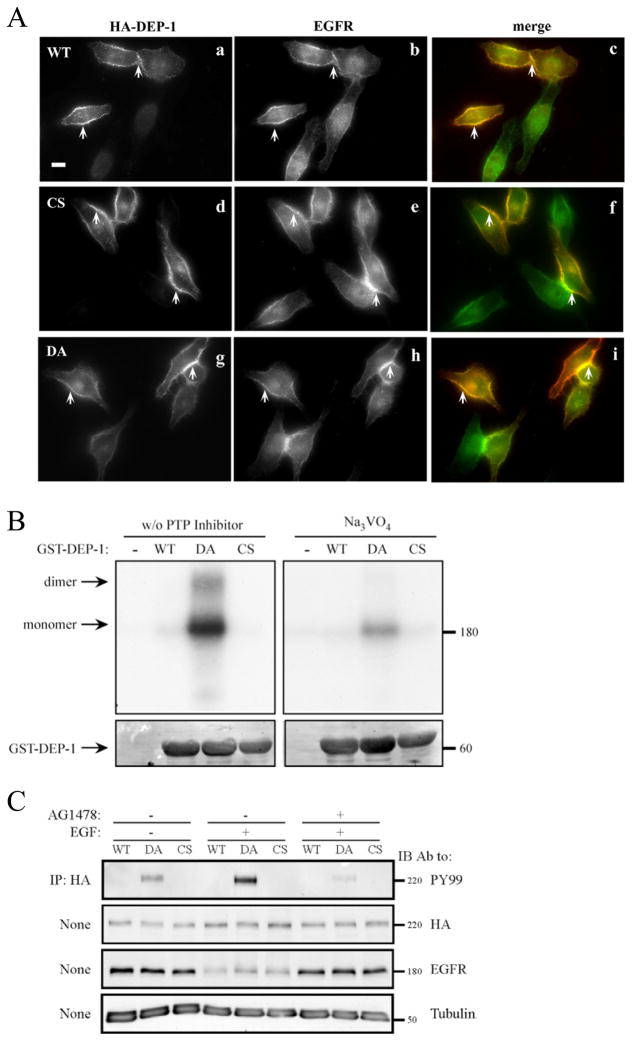
Figure 4. DEP-1 and EGFR Co-localize at the Cell Surface, Physically Interact and Maintain Bi-directional Enzymatic Interactions.
(A) HeLa cells were transiently transfected with vectors encoding HA-tagged WT DEP-1 (a-c), a CS mutant (d-f), or with a DA mutant (g-i). Thereafter, cells were incubated for 32h, serum-starved for 16h, and fixed. Shown are immunofluorescence images obtained with the indicated antibodies. The merged images (c, f and i) were obtained through ImageJ Stack RGB Merge plugin and indicate co-localization (yellow) of EGFR (green) and DEP-1 (red). Arrows demarcate co-localization at cell borders and junctions. One representative experiments is shown (n=3). Scale bar: 10 μm.
(B) A431 cells were incubated for 2hrs at 4°C with a radio-labeled EGF (20 ng/ml), then washed and subjected to covalent cross-linking with BS3 (1mM). The cross-linking reaction was subsequently quenched. Cell lysates were mixed, in the absence or presence of sodium orthovanadate (0.2mM), with glutathione beads bound to purified GST, GST-DEP-1 (WT), the DA or CS mutants, and incubated for 12h at 4°C. After extensive washing, the samples were resolved by gel electrophoresis and autoradiography (top panel). Staining with Ponceau Red (bottom panel) was used to verify equal gel loading.
(C) HeLa cells were transfected with vectors encoding HA-tagged WT DEP-1, or with the CS or DA mutants. Thereafter, cells were incubated for 32h, serum-starved for 16h, and then pre-incubated (as indicated) for 30 min with a selective EGFR kinase inhibitor (AG1478; 10 μg/ml). This was followed by EGF stimulation (20ng/ml; 10 min). DEP-1 was immunoprecipitated (IP) from whole cell lysates using anti-HA-agarose beads, followed by immunoblotting (IB) with the indicated antibodies.