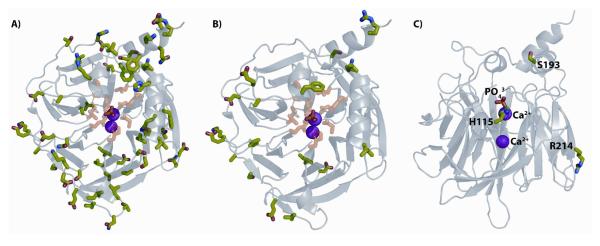
Figure 1. Structure of G2E6 PON1.
A) Residues that differ between human and G2E6 PON1 are rendered as sticks in green. The 59 amino acid differences are scattered throughout the sequence, mainly on the surface of the enzyme and not in the presumed active site. Residues with atoms within 5 Å of the phosphate ion in the crystal are rendered as sticks in pink. The purple spheres represent the calcium (Ca2+) ions. B) Residues that differ between G3C9 and G2E6 PON1 are rendered as sticks in green, where residues with atoms within 5 Å of the phosphate ion in the crystal are rendered as sticks in pink, as in 1A. C) Residue H115 is proximal to the ‘catalytic’ Ca2+ and phosphate ion found in the crystal. S193 lies at the top of the ‘lid’ that is proposed to be an HDL binding site. R214 is on the surface. Images are rendered with PyMOL (DeLano Scientific) from PDB entry 1V04.