Abstract
Glial cells are responsible for a wide range of functions in the nervous system of vertebrates. The myelinated nervous systems of extant elasmobranchs have the longest independent history of all gnathostomes. Much is known about the development of glia in other jawed vertebrates, but research in elasmobranchs is just beginning to reveal the mechanisms guiding neurodevelopment. This study examines the development of glial cells in the bamboo shark, Chiloscyllium punctatum, by identifying the expression pattern of several classic glial and myelin proteins. We show for the first time that glial development in the bamboo shark (Ch. punctamum) embryo follows closely the one observed in other vertebrates and that neural development seems to proceed at a faster rate in the PNS than in the CNS. In addition, we observed more myelinated tracts in the PNS than in the CNS, and as early as stage 32, suggesting that the ontogeny of myelin in sharks is closer to osteichthyans than agnathans.
Keywords: shark, glia, GFAP, myelin, glia development
1. Results and Discussion
Glial cells are arguably the most versatile and fascinating cells in the nervous system. They are responsible for a wide range of functions including general functions like providing anatomical and nutritional support, and fundamental functions such as myelination and scaffolding paths in corticogenesis in the nervous systems of organisms (Jacobson, 1991; Rakic, 1988). All glial cells in vertebrates originate from neuroectodermal tissue (Jacobson and Rao, 2005). Central nervous system (CNS) glia appears during development from dividing cells in different parts of the neural tube: intermediate, marginal and ventricular zones and from radial glia themselves (Rakic, 1971). Peripheral nervous system (PNS) glia are mainly neural crest-derived, with crest cells differentiating into glial cell precursors either during active migration or once they have reached their final destination (Jessen and Mirsky, 2005; Ndubaku and de Bellard, 2008).
Glia in amniotes are known to originate in two main waves (Kettenmann and Ransom, 2005). The first emergence of glial cells occurs during the peak of neuronal development, and those cells are referred to as radial or ependymal glia; they generally disappear in mammals after the neurons have migrated along their processes to reach their final brain/spinal cord layer (Jacobson, 1991; Levitt et al., 1983; Schmechel and Rakic, 1979). The second emergence occurs later in development, and consists of astrocytes, oligodendrocytes, microglia and Schwann cells; these cells will remain within the nervous system throughout life. Thus, during the course of embryonic development radial glial cells are found early and astrocytes and oligodendrocytes are found later. The PNS glia, Schwann and satellite cells, have a special origin; they are neural crest derived: a group of cells that delaminate early in development from the dorsal neural tube, migrating extensively and giving rise to the PNS (Ndubaku and de Bellard, 2008; Trainor et al., 2003).
The cartilaginous fishes belong to one of two extant lineages of gnathostomes (jawed vertebrates). The comparison between their developmental genetic mechanisms and those in the sister lineage, osteichthyans (bony fishes) can provide essential insight into the nature of developmental events in the common ancestor of jawed vertebrates (Venkatesh et al., 2006; Venkatesh et al., 2007). The cartilaginous fishes are the descendants of one of the oldest surviving vertebrate groups, 400 million years ago (Cole and Currie, 2007). Importantly, this group shares a fully myelinated central and peripheral nervous systems with their sister group, the osteichthyans (Bakay and Lee, 1966; Waehneldt, 1990) and all other gnathostomes. Compact myelin is accomplished by glial cells when they tightly ensheath their plasma membrane around an axon, providing insulation and increasing speed of nerve conduction. This structure arose as a gnathostome trait (Schweigreiter et al., 2006; Zalc, 2006) as agnathans do not have compact myelin, but only a single layer of glial cell membranes around their larger axons (Waehneldt et al., 1987). In addition to this distinction regarding myelination, gnathostomes are different from agnathans in that they do have myelinating oligodendrocytes in their CNS while agnathans do not have oligodendrocytes (Rovainen, 1979). Therefore, glial cell development is significantly different between gnathostomes and agnathans.
In the late 1800s, elasmobranchs and especially sharks, held a pre-eminent position in the field of comparative embryology, but in the second half of the 20th century they were replaced by the use of more culture-friendly organisms such as chickens, frogs, mice and zebrafish (Balfour, 1874; Balfour, 1880; Cole and Currie, 2007). However, in recent years a more detailed analysis of neurodevelopment in shark embryos is gaining value as new techniques such as orthologous gene identification and in situ hybridization are re-invigorating their use as models for evolution of development. Hence, the few molecular studies on the early development of shark nervous system are all quite recent. A good number of these have looked at the expression of some key ortholog transcription factors like Otx (Sauka-Spengler et al., 2001), Pax, NeuroD and Phox2B (Derobert et al., 2002; O'Neill et al., 2007) and FoxD (Wotton et al., 2008). These studies illustrate that the formation of the shark nervous system follows a pattern that is highly conserved among agnathans and gnathostomes, and the roles of the described transcription factors in brain regionalization have been highly conserved during vertebrate evolution (Derobert et al., 2002). These studies generally focused on neuronal and placodal development at stages 17–29, between the end of gastrulation and advanced organogenesis (Kuratani and Horigome, 2000). Gould and co-workers examined neural markers like O1, O4, GFAP and neurofilament in Squalus acanthias (dogfish) embryos on a 9cm pre-hatching embryo (Gould et al., 1995), and demonstrated that the same relationships between oligodendrocytes and axons exist during early stages of myelination (Schweigreiter et al., 2006). In this study, we provide further insight into the appearance of glial cells in shark embryos before pre-hatching stages of development (stages 25–29 than in existing literature.
For glial development examination we chose two of the most widely used glial markers, glial fibrillary acidic protein (GFAP) and S100. GFAP, a member of the intermediate filament family, is known for its role in providing strength and support to cells (Kaneko and Sueoka, 1993). Specifically, GFAP forms the intermediate filaments that are characteristic of astrocytes and radial glia to regulate their shape and motility (Kaneko and Sueoka, 1993). This cytoskeletal component has a long phylogenetic history as it has immunoreactivity in the nervous systems of hagfish, lungfish, annelids and mollusks (Cardone and Roots, 1990; Dahl et al., 1985; Lazzari and Franceschini, 2004; Onteniente et al., 1983). The other glial marker we used was S100, a member of multi-member low-weight protein family with a variety of extracellular and intracellular functions, such as regulation of protein phosphorylation, calcium homeostasis, cell growth and differentiation, inflammatory responses, and transcription factor regulation (Donato, 2003; Riuzzi et al., 2006). Past research has demonstrated that GFAP (Ari and Kalman, 2008a; Ari and Kalman, 2008b; Kalman and Gould, 2001) and S100 (Ari and Kalman, 2008a; Chiba, 2000) clearly label glial cells in elasmobranchs making them ideal candidates to use for studies of glial cell development. However, these studies were performed on adult tissues and not embryonic tissues. Our study will delineate the embryonic development and distribution of glial cells in elasmobranchs.
1.1 The expression of gliogenic and neurogenic markers during central and peripheral nervous system development in shark
1.1.1 Stage 25 shark embryos
In order to begin assessing the presence of glial cells in shark embryos, we looked as early as stage 25 (1.3cm).
CNS
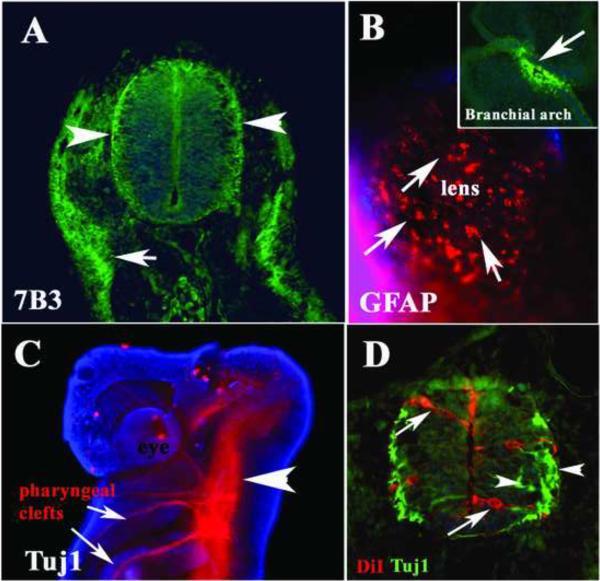
The first glial cells that appear during development in gnathostomes are radial glia, the best known and most abundant type of glia found in developing vertebrates. This glial type is usually distinguished by expressing both nestin and GFAP (Malatesta et al., 2003). In stage 25 shark embryos we observed only weak staining using a polyclonal GFAP antibody (data not shown). In addition, we tested the glial precursor antibody, 7B3, against the transitin protein, also known to label neural crest cells that will later give rise to Schwann cells (Henion et al., 2000) and found that as GFAP, it weakly stained cells in the neural tube (Fig.1A). The GFAP and 7B3 staining was observed in both the ventricular and marginal layers of the developing neural tube. Interestingly, on the ectoderm/skin of these shark embryos there were scattered cells clearly positive for GFAP (arrows in Fig.1B). Although we did not observe strong staining for GFAP or 7B3 in the neural tubes at this stage, we observed neuroblasts labeled with DiI extending along the width of the neural tube (Fig.1D). We also observed that cells in the marginal zone were labeled with anti-beta III neuronal specific tubulin, Tuj1 (arrowheads in Fig.1C, D) indicating that neurogenesis is actively taking place at this stage.
Figure 1. Neural markers expressed in the Stage 25 shark embryos.
Immunofluorescence of st.25 shark embryo for 7B3/transitin (A) GFAP (B) and Tuj1 (C, D). A Thin section through one embryo at the mid-trunk level shows positive 7B3 cells in the neural tube, especially on the ventricular and laminar side of the neural tube (arrowheads) and developing lateral line underneath the ectoderm (arrow). B Ectoderm around the eye shows cells positive for GFAP (arrows). Head (C) and trunk (D) immunostaining with the tubulin-III neuron specific antibody Tuj1 showed positive neural tube and axonal projections into the branchial arches (C). Cell nuclei were stained blue with DAPI.
PNS
At stage 25, shark embryos have extensive CNS growing axons into the pharyngeal clefts (or branchial arches, Fig.1C) and in the forming spinal nerves along the trunk (data not shown). More importantly, cells migrating on the branchial arches (presumptive neural crest cells) were positive for GFAP (insert in Fig.1B) and the pre-migratory neural crest also weakly expressed GFAP (data not shown). We found positive staining for 7B3 along what is likely to be migrating lateral line primordium (arrow in Fig.1A).
1.1.2 Stage 27 shark embryos
At stage 27, shark embryos are still growing at the tail by adding new somites.
CNS
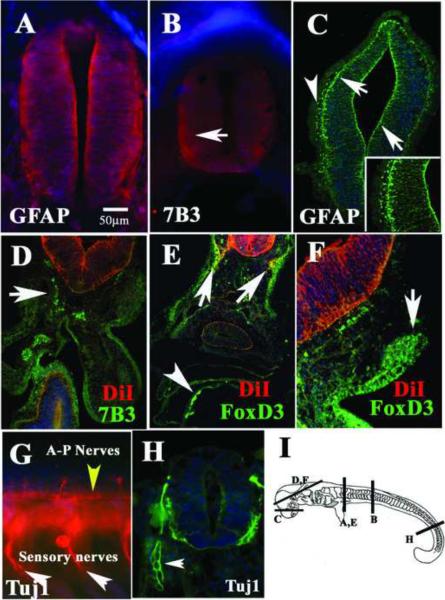
Figure 2A shows stage 27 shark embryos with robust expression of GFAP in the neural tube and a pattern reminiscent of radial glia, especially at the most rostral levels. Myoblasts stained for GFAP as well (data not shown). Robust expression of 7B3 was seen in rostral neural tube cells extending throughout the neural tube (Fig.2B) as was also shown for GFAP (Fig. 2A). Staining of thin sections with GFAP antibody in the forebrain showed a clear radial pattern of fibers extending throughout the width of the neural tube (arrows in Fig.2C). GFAP and 7B3 staining at more caudal, less developed neural tubes were weak, as was seen in stage 25 embryos (data not shown).
Figure 2. Neural markers expressed in the Stage 27 shark embryos.
Immunofluorescence of st.27 shark embryo for GFAP (A, C), 7B3 (B, D), FoxD3 (E, F) and Tuj1 (G, H). By this stage neural tube shows robust staining for glia markers (GFAP in A and C, 7B3 in B). GFAP was expressed in radial glia (arrows in C) and neural crest (arrowhead in C). 7B3 and FoxD3 labeled migrating neural crest (D), lateral line (E) and cranial ganglia (F). The neuronal marker Tuj1 was widely expressed by peripheral nerves (G, H) and by ventricular cells in the neural tube (H). I shows a cartoon of a stage 27 indicating the regions of the sections in each figure. D–F neural tubes were vitally labeled with DiI, shown in red channel. Cell nuclei were stained blue with DAPI.
PNS
GFAP surprisingly stained what could be cranial migrating neural crest cells (arrowhead in Fig.2C) and 7B3 labeled mesenchymal cells in the shark embryos along the migratory pathways of neural crest cells (Fig.2D). To identify the 7B3-positive mesenchymal cells as putative neural crest/glial cells we used FoxD3, a transcription factor that labels pre-migratory and early migrating neural crest in zebrafish (Lister et al., 2006). FoxD3 antibody strongly labeled DiI migrated neural crest along lateral line region (arrows in Fig.2E), thus supporting the hypothesis that these cells are putative peripheral crest derived glia. FoxD3 also labeled the ectoderm in shark embryos in a similar cellular pattern to that of ectoderm GFAP-expressing cells at stage 25 (data not shown) and what appears to be a developing cranial ganglia (arrow in Fig.2F).
Wholemount staining with the neuronal marker Tuj1 at stage 27 showed a robust peripheral neuronal development. At the most rostral levels, there are already clearly defined spinal nerves and axons growing anterior to posterior in the embryo (yellow arrowhead in Fig.2G). At the most caudal levels, where the nervous system of the embryo is still in early phases of development, thin section showed that the cells on the laminar layer as well as developing dorsal root ganglion express Tuj1 (Fig.2H). However, observed weak GFAP-positive staining in these peripheral nerves at this stage (data not shown). In both wholemount and thin section, the Tuj1 immunostaining showed that peripheral ganglia have more Tuj1 positive cells than neural tubes (arrowhead in Fig.2H).
1.1.3 Stage 29 shark embryos
In stage 29 embryos, the nervous system growth (PNS and CNS) is more advanced and the embryo is starting to change towards a more shark-like shape.
CNS
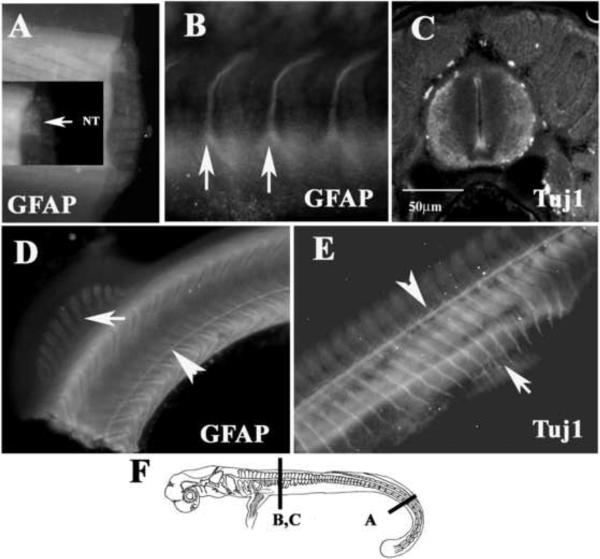
Embryo staining with GFAP showed that there are enough differentiated glial cells in the CNS at stage 29 to highlight the whole spinal cord in wholemount staining in a diffuse manner (Fig.3A). At more rostral levels, the laminar part of the neural tube was positive for Tuj1 (Fig.3C).
Figure 3. Neural markers expressed in the Stage 29 shark embryos.
Immunostaining staining for GFAP (A, B, D), Tuj1 (C, E) and HNK-1 (F-H), in a st.29 embryo. GFAP stained neural tube (A) and lateral line (arrowhead in D), spinal nerves (arrows in B) and migrating myoblasts (arrow in D). Lateral line (arrowhead in E) and spinal nerves (arrow in E) are also positive for Tuj1. F show a cartoon of a stage 29 embryo with line indicating the level of images in figures.
PNS
At the most rostral level, where the nervous system is more mature, GFAP highlighted a set of peripheral nerves (arrows in Fig.3B). These nerves and lateral line (Fig.3D) are well into differentiation as indicated by Tuj1 wholemount immunofluorescence (arrowhead in Fig.3E). Peripheral nerves are beginning to enter the rostral fin (arrow in Fig.3E). The developing myoblasts in the caudal fin also showed strong GFAP expression (arrow in Fig.3D).
1.1.4 Stage 32 shark embryos
The CNS of shark embryo at stage 32 is well developed, as there are clearly visible neuronal layers in the hindbrain and the PNS seems to have reached its maximal sensory ganglia formation (we did not see DRG at the most caudal levels, only projecting nerves, data not shown). There are many nerves extending throughout the embryo, especially into the muscle tissues.
CNS
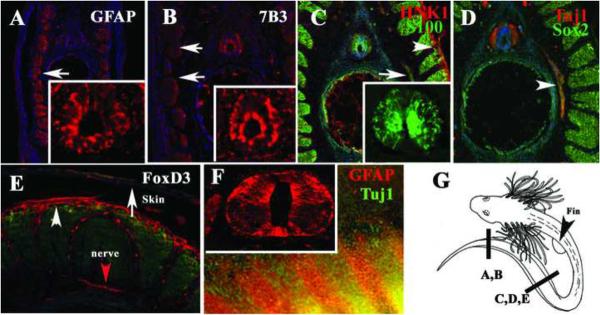
Immunohistochemistry on sections at the tail-most level in stage 32 embryos demonstrated that GFAP is strongly expressed in a radial pattern in the developing spinal cord (Fig.4A) as well as in more rostral sections (insert in Fig.4F). Similarly, 7B3/transitin is expressed in an identical radial pattern in the neural tube (Fig.4B). In addition, we observed that both markers, GFAP and to a lesser degree, 7B3, continued to be present in the developing myoblasts, (arrows in Fig.4A, B). The S100 glial marker was present at this stage in a similar radial pattern as GFAP and 7B3 (Fig.4C). Interestingly, we were not able to determine with certainty the presence of S100-positive cells in the previous stages.
Figure 4. Glial and neural crest markers expressed in the Stage 32 shark embryos.
Sections through a st.32 embryo at the tail most end for GFAP (A), 7B3 (B), S100 (C), Tuj1 (D) and FoxD3 (E) labeled a specific group of cells. Radial glia and myoblasts were strongly stained with GFAP (insert for neural tube magnification in A and F). 7B3 labeled radial cells in the neural tube (insert in B) and to a lesser extent in the medial myoblasts (arrows). C S100 (green) also labeled radial cells; insert shows a magnification of the neural tube, while HNK-1 labeled skin (arrowhead). D Tuj1 (red) and Sox2 (green) weakly overlap and double labeled peripheral nerve (arrowhead in D). E FoxD3 was present in peripheral nerve (red arrow) as well as in dermal cells (arrowhead in E). GFAP labeled a group of cells in the fin (red arrows in F) while Tuj1 presented a banded staining in the fin (green). G shows a cartoon of a stage 32 embryo with line indicating the area for each figure. Cell nuclei were stained blue with DAPI.
PNS
Peripheral nerves in stage 32 embryos were stained with S100 at the trunk level (arrow Fig.4C). At similar antero-posterior level Tuj1 labeled the neuronal component (arrowhead in Fig.4D). The neuronal/stem cell marker Sox2 faintly stained peripheral nerve (arrowhead in Fig.4D), and gave strong positive signal in younger myoblasts (Fig.4D) at more caudal levels, along antero-posterior axis, whereas developmentally older myoblasts, at rostral levels, did not express Sox2 (data not shown).
HNK-1 labeled a group of cells under the skin (arrowhead in Fig.4C) similar to FoxD3. FoxD3, a transcription factor responsible for neural crest and glia development (Kelsh et al., 2000; Kos et al., 2001), was expressed in cells underlying the skin ectoderm (arrowhead in Fig.4E) (Dottori et al., 2001) and most importantly in peripheral nerve (red arrowhead Fig.4E).
GFAP wholemount staining in the trunk showed the same pattern as in stage 29 embryos; positive myoblasts (not shown) and a banded signal in the fin which partly overlapped with the Tuj1- and HNK-1 expression domains (Fig.4F and data not shown).
We tested several commercially available antibodies against the SoxB and SoxE transcription factors (Sox8, Sox10) to see if we could label peripheral glia in shark embryos. But none of the antibodies against proteins expressed in glia (Sox8 and 10) yielded specific staining probably because they do not recognize the most conserved HMG domain in SoxE gene family members (Dutton et al., 2008).
1.2 Pre-hatching shark embryos
Once C. punctatum embryo has reached 6 cm of length, their body morphology resembles that of a small adult bamboo shark. Since development progresses in a rostral to caudal manner, we examined the expression of glial markers at the tail, mid-trunk and cephalic regions. Because at this stage, the rostral and most of the trunk regions of the nervous system are well advanced in development, we expected that the use of the glial markers would provide more structural rather than developmental information.
CNS
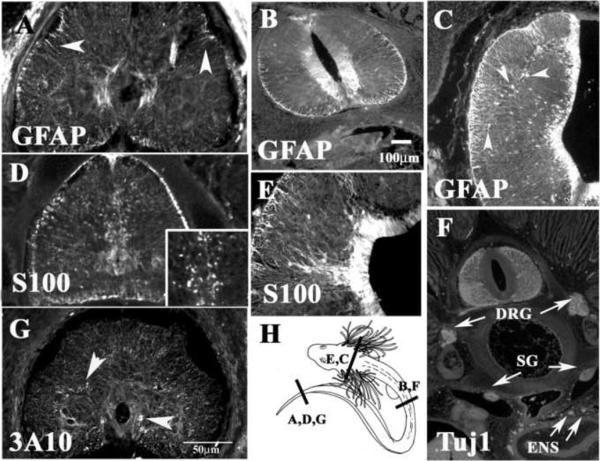
At the most caudal end of these embryos, we detected extensive staining in radial glial fibers using GFAP and S100 antibodies (Fig.5A and D). The expression of these GFAP- and S100-positive cells was prominent around the central canal and in fibers projecting to the periphery of the spinal cord, although none were “star-shaped”, as expected (Ari and Kalman, 2008a). S100 also showed some punctuated staining likely corresponding to glial cells in the ventricular zone of the spinal cord (Wicht et al., 1994) (Fig.5D insert), but we could not definitively identify them as either oligodendrocytes or astrocytes. To determine the level of development of neurons/neuroblasts at this anatomical level, we used the well known neural marker 3A10 that labels neurofilament in neurons in other species of sharks (Freitas and Cohn, 2004; Freitas et al., 2006; O'Neill et al., 2007). We observed that, at the trunk level, positive fibers in the spinal cord labeled the same discreet areas that myelin markers identified, which strongly suggests that they correspond to axons in the process of myelination (Potzner et al., 2007) (see Fig.5G and also Fig.7).
Figure 5. Neural markers expressed in the pre-hatching shark embryos.
Shark embryo sections were immunostained for GFAP (A–C), S100 (D, E), 3A10 (G) and Tuj1 (F). GFAP and S100 both labeled radial glia processes, while S100 also cell bodies in the ventricular zone (magnified insert in D). DRG, sympathetic ganglia (SG), motor axons (Ax) and enteric nervous (ENS) system were all clearly marked with Tuj1 in F.
Figure 7. Myelin markers expressed in the trunk of pre-hatching shark embryo CNS.
Spinal cord sections were immunostained for Tuj1 and MPZ (A–D), CNPase (E), MBP (F, H) and PLP (G). Spinal cord and nerves had abundant neuronal processes as attested by Tuj1 and myelinated fiber are beginning to delineate the butterfly shape of mammalian spinal cords observed in osteichthyans (B, F and to lesser extent E). At the tail-most end, MBP labeled fewer axons along the same locations as in more rostral regions (H). PLP did not give positive staining on these sections (G). Arrows point to myelinated tracts.
The expression pattern of GFAP and S100 at the rostral end of these embryos was quite different. GFAP- and S100-positive fibers were present in abundance in both head and trunk (Fig.5B, C, E). GFAP staining was observed in a classic radial fashion (Dahl et al., 1985) and in great abundance in the ventricular zone along the length of the neural tube, much more pronounced than in stage 32 embryos (Fig.5B–C). The definite presence of star shaped cells are indicative of astrocytes (arrowheads in Fig.5C) (Ari and Kalman, 2008a). S100 staining was very similar to GFAP especially in the head (Fig.5E), and to lesser extend in the spinal cord (data not shown).
PNS
It is clear that by this stage neuronal development is well advanced, since Tuj1 staining was present along the entire neural tube including all the levels in the head and trunk. Figure 5F shows a section at the trunk level stained with Tuj1, where the presence of mature neurons exist not only in the developing spinal cord, but also as motor axons, sympathetic ganglia, and the gut enteric nervous system. At this stage, GFAP levels in nerves was reduced in comparison to other stages and to the CNS (data not shown).
1.3 Study of Myelin markers in the shark embryos
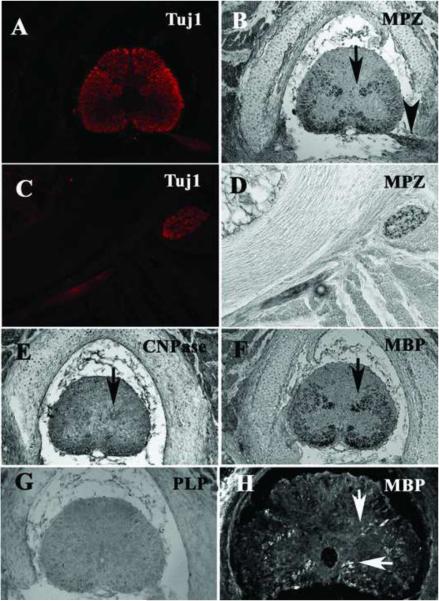
One of the main markers of myelination is myelin basic protein (MBP). This protein is the second most abundant myelin component in vertebrates, and is made by both oligodendrocytes and Schwann cells (Allinquant et al., 1991; Morell, 1984). Immunostaining of rostral sections of stage 32 embryos showed that MBP as well as Myelin Protein Zero (MPZ, Po) is starting to appear in the midbrain of these embryos (Fig.6A and C). In a late stage 32 embryo, but not yet pre-hatching, we observed more specific staining of myelinated nerves using MBP antibody in the peripheral ganglia. Specifically, trigeminal ganglion at cranial level and dorsal root and spinal nerves in the trunk stained positive for MBP (Fig.6B and D), while very little staining was found in CNS.
Figure 6. Myelin markers expressed in brain of shark embryo.
Section staining for MPZ in hindbrain (A, C) in early stage 32 embryo showed that myelination is starting. In later stage 32 embryos MBP (B, D) showed that a cranial ganglion is undergoing extensive myelination (arrows point to myelinated nerves). Pre-hatching tegmental areas showed myelinated fibers crossing the brain (E, F). MPZ labeled also cell bodies in E.
However by pre-hatching embryo stage, myelin tracts positive for MPZ and MBP were distinctly present in the CNS (Fig.6E and F), while few myelinated fibers were present in the brain (data not shown). MPZ was expressed not only in myelinated fibers, but also in the bodies of some cells in the CNS (squared area in Fig.6E).
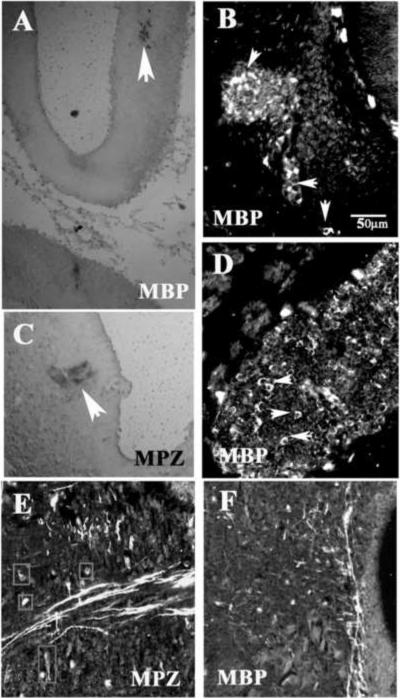
In addition to looking in the head regions, we also looked in the trunk for presence of myelinated tracts in the spinal cord and PNS. Interestingly, in the pre-hatching embryo the levels of myelination in both tissues were more abundant than in the brain as indicated by MPZ, CNPase and MBP specific and strong staining (Fig.7A–F). Neither of these tissues showed PLP expression (Fig.7G). Double staining with Tuj1 and myelination markers showed that the neural tube has the morphology and expression patterns of a developmentally mature spinal cord (Fig.7A) and that both Tuj1 and MPZ labeled identical regions in the spinal cord, namely the sensory ganglia and peripheral nerve (Fig.7C and D). A few clusters of myelinated fibers were positive for MBP at the most caudal portion of the spinal cord (Fig.7H), specifically from the ascending/descending tracts, and mostly in the ventral portion. At this same caudal level, we found that SMP antibody stained fibers in a similar manner (data not shown). These results indicate that the process of myelination progresses faster in the spinal cord than the head.
Indeed, we found that in stage 32 embryo both cranial and peripheral ganglia expressed higher amounts (in pattern, distribution and intensity) of myelin proteins (MPZ, SMP, PLP and MBP) compared to CNS. At the level of spinal cord, MPZ myelin protein was robustly expressed in the DRG (Supplementary Fig.8A) and spinal nerve (not shown). MBP expression was always more robust than MPZ. However, not all the peripheral axons were myelinated at this stage as shown by only half the ciliary ganglion staining for MBP (Supplementary Fig.8C) and only part of the trigeminal staining for SMP (Supplementary Fig.8D). PLP was strongly expressed in the VIII cranial ganglia (Fig.8B), though we did not observe significant staining in the spinal cord. More importantly, the pattern for PLP staining indicated more cell surface staining of neurons (as for p75 and Tuj1 in Supplementary Fig.8E and F) than of myelinated tracts as was observed for MBP, MPZ and SMP.
1.4 Glial formation
The vast majority of studies on chondrichthyan glia have been on adult or pre-hatching embryo spiny dogfish (Squalus acanthias), a species phylogenetically related to bamboo shark, or skate species (Ari and Kalman, 2008a; Gould, 1992). These studies focused on the presence and distribution of astrocytes and radial glia in mature brains. Our findings showed that the first detectable glia appear between stage 25 and 27, using GFAP and 7B3 antibodies for identification. In stage 25 embryos, we observed 7B3 and GFAP in cells whose morphology suggests neuroepithelial cells instead of the radial cell as was observed in zebrafish embryos, further confirmed by live DiI labeling (Kim et al., 2008). By stage 27, GFAP and 7B3-positive cells spanned the rostral neural tube, and their morphology suggested that their phenotype corresponds to true radial glia instead of neuroepithelial (Malatesta et al., 2003). These cells have been already described in adult agnathans like lamprey and in adult sarcopterygians like lungfish (the sister group to the tetrapods), as well as in osteichthyans and actinopterygians embryos (Campbell and Gotz, 2002; Park et al., 2004; Rakic, 1971) based on being positive for glial markers (see Table II). While radial glia appearance in mouse is known to happen around embryonic day 12 (Hartfuss et al., 2001), shortly followed by neurogenesis, these cells soon after will disappear (Cai et al., 2005; Malatesta et al., 2003). In agnathans and chondrichthyans, these cells will remain through adulthood (Rovainen, 1979). The early appearance of radial glia during neurogenesis indicates a similarity in neural development between chondrichthyans and amniotes that does not seem to be shared with actinopterygians.
Table II.
Summary Table of Glia development in some vertebrates
| Organism | Earliest Glia found | Markers Used | References | ||
|---|---|---|---|---|---|
| Agnathan | Lamprey (Lampreta japonica) | Adult lamprey: GFAP immunoreactivity observed in long straight processes that radiated through neurophil from ventricular wall, following nerve fiber bundles in white matter. | GFAP | (Onteniente et al., 1983) | |
| Chondrichth yan | Shark (Squalus acanthias and Chiloscyllium punctatum) |
Pre-hatching embryo: 5.5 cm (myelin) 9 cm (Ast, Ols) |
Po,MBP,O1,O4 | (Gould et al., 1995; Gould, 1992;Rotenstein et al., 2008) | |
| Osteichthyan | Actinopterygian | Fish (Danio Rerio) |
Embryo: Radial glia show at 24hpf Embryo: Schwann cell show at 24hpf |
GFAP-GFP promoter Fkd6 | (Kim et al., 2008) (Kelsh et al., 2000) |
| Sarcopterygian | Coelacanth (Latimeria chalumnae.) | Adult: glia observation was from myelin studies. | Myelin proteins (PLP/DM20) | (Linington and Waehneldt, 1990; Tohyama et al., 1999; Waehneldt and Malotka, 1989) | |
| Lungfish (Protopterus annectens) | Adult lungfish. GFAP: immunopositive structures represented by thin long fibers of cells were found in both grey and white matter of CNS, as well as thin fibers that converged on blood vessel walls. Vimentin immnoreactivity was observed in peripheral zone of brain, but weakly observed in the white and grey matter fibers of the brain and the white matter of the ventral region of spinal cord. | GFAP and vimentin | (Lazzari and Franceschini, 2004) | ||
| Amphibian (Xenopus laevis) | Stage 21: Brain: NF expression in developing neurons occasionally observed at stage 21 and reliably in the neural tube and caudal regions of the brain at st 23. At stage 23, GFAP expression was observed in the cytoplasm of spinal cord radial glial but only in the peripheral region of the brain. | GFAP and NF | (Messenger and Warner, 1989) | ||
| Osteichthyan | Sarcopterygian |
Reptiles (Eumeces fasciatus) (Gallotia galloti) Turtle (T.sinensis) |
E32-juvenile (Ols) Stage 29-CNS myelination Stage 32: Telencephalon: Vimentin observed in the proliferative zone of the lateral ventricle and subpial end-feet in the marginal zone at stage 32. At stage 34–35, vimentin staining is at its peak of intensity in all radial glial. GFAP appears at stage 35 in the end-feet in the marginal zone and increases until adulthood. 5–6 cm-juvenile (ependymal glial cell pattern throughout CNS; extra-ependymal star-shaped astrocytes in the spinal cord) |
S100, Electron microscopy Vimentin and GFAP GFAP No vimentin immunoreactivity was observed |
(Monzon-Mayor et al., 1990; Romero-Aleman et al., 2004). (Nadon et al., 1995) (Romero-Aleman et al., 2004) (Lazzari and Franceschini, 2006) |
| Birds Chicken (Gallus gallus) |
Radial Glia. Observed as early as E5 using recombinant virus. Astrocytes: Observed as early as E10 Oligodendrocytes: precursors are early, but they have been observed by: E6 using Nkx2.2-positive cells E12 using transplant and SMP antibody Schwann cells: seen in neural crest population by HH19. ckE14: Schwann cells precursors are detected with MPZ. |
Recombinant virus. CD44 Nkx2.2 SMP Seraf 1E8, S100, O4 |
(Gray and Sanes, 1992) (Alfei et al., 1999) (Fu et al., 2003) (Cameron-Curry, 1995) (Wakamatsu et al., 2004) (Bhattacharyya et al., 1991) |
||
| Osteichthyan | Sarcopterygian |
Mammals Rodents (Rattus rattus and Mus musculus) Primates (Macacus Rhesus monkey) |
Radial Glia. GFAP promoter directed expression in RG by E12. RC1 expression seen at E13.5–14.5RCl+ cells were nestin immunoreactive at E18.5. RC1 expression partially overlapped with GFAP at this stage. Astrocytes: GFAP expression detected first at E16. It was weakly expressed in dev axonal tracts. CD44 was detected as early as E11.5 in rat spinal cord. Oligodendrocytes: in rat spinal cord by E12. Schwann cells: E11 Radial glial appear in first third of gestation. They appear in brain and spinal cord by E41; in the diencephalon by E45; and in the telencephalon and cerebellum by E47. Bergmann glial cell of the cerebellar cortex appear by E54 |
RC1, GFAP, vimentin, nestin, CD44 (possible astrocyte marker). PLP-GFP promoter Rat Po mRNA in situ |
(Liu et al., 2002; Malatesta et al., 2003) (Barry and McDermott, 2005; Liu et al., 2002) (Le Bras et al., 2005) (Lee et al., 1997; Woodhoo et al., 2004) (Levitt and Rakic, 1980) |
Ast: astrocyte
Ols: oligodendrocyte
SC: Schwann cell
RG: radial glia
The expression of GFAP in pre-hatching embryos in the bamboo shark (Ch. Punctatum) was very strong along the ventricular layer and in the numerous radial fibers present throughout the CNS as has been observed in other vertebrates (Csillag and Kalman, 2008; Kalman, 2002; Kalman and Gould, 2001). This pattern of GFAP expression (and S100) in the pre-hatching embryo was more like the one present in osteichthyans and in hagfish (Wicht et al., 1994) but it did not look like the one we found in earlier embryos when using DiI. This difference in shape and timing of cell appearance suggests there may be two different types of radial glia in shark embryos. An early radial glia along which neurons climb like those in osteichthyans, and a later radial glia that remains in the embryo through adulthood like those thought to be stem cells in zebrafish (Kim et al., 2008).
There are few markers that indicate the presence of immature/precursors of glial cells: for example A2B5, FoxD3, Krox20, Nkx2.2 and Sox10 (Kettenmann and Ransom, 2005; Ndubaku and de Bellard, 2008). We successfully achieved positive signal from only two antibodies we tested : FoxD3 and 7B3/transitin. Suggesting that both these transcription factors, or their close paralogs, are present in the bamboo shark (Wotton et al., 2008). Recently, Appel and co-workers demonstrated the existence of ventrally migrating glia in zebrafish that give rise to nerve perineurium (Kucenas et al., 2008). Unfortunately, even though the presence of these ventrally migrating glia have been shown in lamprey, chicken and zebrafish (Lunn et al., 1987; Nakao and Ishizawa, 1987), our use of Nkx2.2 gave ambiguous results for the presence of these cells in bamboo shark.
The appearance of astrocytes must be a later phenomenon in bamboo shark since no star-like shaped cells were observed in our stage 27 through 32 embryos, though they were clearly present on the pre-hatching embryos. Astrocytes appear late in development in mammals, reptiles and birds (Kalman, 2002; Sanosaka et al., 2008; Woodruff et al., 2001; Wu et al., 2006). The bamboo shark may follow the osteichthyan program for astrocyte development than the one found in agnathans (hagfish and lamprey). Past research from Kalman's lab and others showed the presence of astrocytes across several species of adult elasmobranchs and that the differences among them were mainly in their abundance throughout brain regions (Ari and Kalman, 2008a; Ari and Kalman, 2008b; Kalman and Gould, 2001). Our study did not look into specific conserved brain regions but focused on early shark embryos. Future studies using Glutamine synthetase (GS) and vimentin antibodies in stage 32 onwards may provide more detailed information about shark astrocytes development and origins (Ari and Kalman, 2008a; Ari and Kalman, 2008b).
It has been documented that glial cells and myelinated tracts of the lizard Gallotia galloti, have S100 immunoreactivity (Romero-Aleman Mdel et al., 2003) and that mature astrocytes and tanycites in juvenile dogfish, Scyliorhinuscanicula. express S100 (Chiba, 2000). However, no studies have concentrated on the earliest stages of shark embryogenesis using this marker. Our study shows for the first time the presence of S100 in the developing nervous system of the shark, especially its presence in the radial glia of the developing neural tube and in peripheral nerves. The S100-positive cell bodies found in the spinal cord, most likely correspond to oligodendrocyte precursors as suggested by the overlap of S100 antibody staining and in situ hybridization with a probe specific for shark Sox8 (manuscript in preparation).
1.5 Myelin development
A very important aspect in the evolution of nervous system development of gnathostomes is the appearance of myelinated axons (Zalc, 2006). The ontogenesis of myelin has been a controversial issue. Some studies show that ontogenically myelin markers appear first in the PNS before CNS of fish, mice and chicken (Brosamle and Halpern, 2002; Fu et al., 2002; Martenson, 1992; Wang et al., 2006) although it is not settled if this pattern (first PNS then CNS) also happens phylogenetically. Other studies suggest that myelin first appears in the CNS, or simultaneously between the CNS and the PNS (Zalc, 2006). In the meantime, no definitive ontology has arisen on the development of myelin in sharks. Previous studies indicated that ventral spinal cord of pre-hatching dogfish (Squalus acanthias) have myelinated tracts (Gould, 1992) and that myelin MPZ and MBP proteins can be found in other shark species in pre-hatching embryos (Rotenstein et al., 2008). Our present study shows for the first time that bamboo shark embryos have myelinated fibers from stage 32 onwards, well before the pre-hatching stage, and that its appearance takes place in the PNS before the CNS. Therefore placing the ontogeny of myelination in sharks along that of fish, birds and mammals, which show myelin markers first in the PNS before the CNS.
Vertebrate myelin sheaths express either MPZ or PLP, usually accompanied by DM20 (Martenson, 1992; Snipes et al., 1993). While MPZ is found in the CNS myelin in fish and shark, PLP/DM20 is generally accepted as the myelin CNS marker in tetrapods, suggesting a switch from a MPZ based myelin CNS to a PLP one in these organisms (Sinoway et al., 1994; Yoshida and Colman, 1996). Our study found as expected from these past studies that shark embryos have much less PLP expression in myelin tracts compared with other myelin proteins (MPZ, MBP and CNPase).
We found that in addition to MBP in early spinal cord, SMP is present in CNS glia as well as in PNS ganglia. SMP is a protein used to differentiate myelin-producing glial cells from glial satellite cells (Cameron-Curry et al., 1991; Dulac et al., 1988) and is detectable in late neural crest-derived cells days before myelination begins (Dupin et al., 1990). Hence, it seems that in chondrichthyans not only MPZ protein, but also SMP is expressed both in PNS and CNS tissues,
In conclusion, our results show for the first time that glial development in the bamboo shark (Ch. punctamum) embryo follows closely the one observed in other vertebrates and that neural development seems to proceed at a faster rate in the PNS than in the CNS. We observed that glial markers were present in shark nervous system starting by stage 25, with radial glia present by stage 27. Myelination was found as early as stage 32 embryos and was more abundant in the PNS than CNS in these shark embryos suggesting that myelin ontogeny in sharks follows the same pattern as in osteichthyans rather than agnathans. Altogether, these data reinforce the relevance of using sharks as model organisms (Coolen et al., 2009).
2. Experimental procedures
2.1 Collection and Staging of Embryos
Bamboo shark cases, Chiloscyllium punctatum (Müller and Henle, 1838) or mermaid's purses were harvested from the Long Beach Aquarium and Cabrillo Aquarium (kindly provided by Chris Plante and Kiersten Darrow, respectively), reared at 25°C in sea water, and collected at different developmental stages. Embryos were removed from egg cases and fixed in 4% Paraformaldehyde or Carnoy's fixative overnight at 4°C. The embryos were staged according to Ballard's developmental table when feasible or by their length in cm (Ballard, 1993). The youngest embryos collected were 3cm-long and had accomplished the development of most of their forebrain. The oldest embryos were 10cm-long, a stage at which they show much physical activity and look like their adult counterparts. Embryos were either fixed in Carnoy's (70% ethanol, 20% formaldehyde and 10% glacial acetic acid) or in 4% paraformaldehyde overnight at 4°C and kept in 70% ethanol at −20°C until histological preparation. For sufficient paraffin penetration, embryos needed very extensive dehydration steps (about 1 day per alcohol grade) and two full days in histosol for clearing. The tissues were then immersed in hot paraffin (McCormick Scientific Paraplast Plus) in a vacuum oven for two days before preparing the blocks and sectioning. Embryos were sectioned (7–12μm) with a microtome, collected on Super-Frost slides and dried overnight at 37°C on a slide warmer. DiI injected embryos were cryoprotected in 15% and then 30% sucrose overnight, embedded in gelatin for 3hrs at 38 °C, slowly frozen in liquid nitrogen, and sectioned at 12μm.
For live labeling, stage 25 and 27 shark embryos were immobilized with tricaine and injected with DiI (cell tracker CM-DiI, C-7001, Invitrogen/Molecular Probes) (diluted in ethanol (1/10) in 10% sucrose) inside the neural tube along its length and hindbrain regions. The embryos were placed in a Petri dish after rinsing in sterile sea water and incubated with 5ml of DMEM, 10% FBS, penicillin and streptomycin at 37°C for 12hrs. These embryos survived for this period of time. At the end of incubation embryos were fixed for 1hr without transplanting them in 4% PFA, then placed in a vial for overnight fixing at 4°C and remained there until cryo-sectioning.
2.2 Immunohistochemistry on tissue sections
Shark tissue sections were de-waxed in histosol and re-hydrated in a graded series of ethanol washes (histosol, 100, 90, 70, 50 and 25% ethanol washes in water) and then equilibrated in PBS (Dulbecco's) before blocking in PBS containing 10% fetal bovine serum (FBS) and 1% Triton X-100 for 1hr at room temperature. Primary antibodies were added in a 1:500 dilution in PBS and slides were incubated overnight at 4°C. A battery of primary antibodies (see Table I) was used at concentrations ranging from 1:100 to 1:500. After washing the sections in PBS 3 times for at least 5 minutes each, secondary antibodies (Alexa fluoroprobes conjugated to anti-rabbit or anti-mouse IgG, Invitrogen) were added for 30 min with DAPI to label the nuclei, washed in PBS for immuno-fluorescence visualization and coverslipped with Permount. In some instances we did DAB staining of embryos or sections (following the protocol from Innovex Technologies, Santa Clarita, CA). Pictures of sections were taken using Axiovision LE software (Zeiss™) with an AxioCam black and white camera attached to a Zeiss AxioimagerA1 upright fluorescent microscope and assembled into figures using Adobe Photoshop 7.
Table I.
Summary Table of antibody results
| Glial | St. 25 | St. 27 | St. 29 | St. 32 | Pre-hatching |
|---|---|---|---|---|---|
| GFAP (cyto) | + (CNS) | + (NS, Plac) | + (NS, myoblasts) | ++ (NS, myoblasts) | +++ (NS) |
| S100 | - | - | - | ++ (CNS, myoblasts) | +++ (CNS) |
| MBP (myelin) | - | - | - | + (PNS) | +++ (NS) |
| MPZ (myelin) | - | - | - | + (PNS) | +++ (NS) |
| SMP (myelin) | - | + (CNS) | +++ (NS) | ||
| CNPase (myelin) | - | - | + (NS) | ||
| PLP (myelin) | - | - | ++ (PNS) | ||
| 7B3/transitin (TF) | +(NS) | + (NS) | ++ (NS) | ++ (NS) | |
| FoxD3 (TF) | + | + | + (NS) | ++ (NS, skin Plac) |
| Neuronal | St. 25 | St. 27 | St. 29 | St. 32 | Pre-hatching |
|---|---|---|---|---|---|
| Tuj1 (cyto) | ++ | ++ | +++ | ++++ (CNS, myoblasts) | ++++ (NS) |
| 3A10 (cyto) | ++ | ++ | ++ (CNS) | ||
| Nf-M (cyto) | ++ | ++ | ++ (NS) | ||
| Sox2 (TF) | ++ (skin Plac) | ++ (Plac) | |||
| p75 (receptor) | - | ++(PNS) | |||
| HNK-1 (carb) | + (Plac) | ++ (Plac) | ++ (CNS, skin Plac,) | ++ (CNS, Plac) |
+, ++, +++ or ++++ = refers to the abundance/intensity of the staining with the antibodies.
- = We did not observe any staining.
Empty (not tested).
CNS (central nervous system), NS (nervous system), PNS (peripheral nervous system). GFAP (glial fibrillary acidic protein), S100 (Calcium binding protein), MBP (myelin basic protein), MPZ (myelin P zero protein), SMP (Schwann cell myelin protein), CNPase (2′, 3′-cyclic nucleotide 3′-phosphodiesterase), PLP (proteolipid protein), 7B3 (transiting), FoxD3 (forkhead family of transcription factors),Tuj1 (anti-beta III tubulin), 3A10 (Neurofilament), NfM (neurofilament medium), Sox2 (SRY 2 transcription factor), p75 (NGF receptor), HNK-1 (antibody recognizes carbohydrate epitope).
Cyto (cytoskeletal protein), TF (transcription factor) and carb (carbohydrate)
Antibodies used were: polyclonal GFAP and S100 (DAKO), monoclonal TuJ1 (Sigma), polyclonal FoxD3 (courtesy of D. Raible (Lister et al., 2006), polyclonal MBP and MPZ raised against shark myelin peptides (courtesy of B. Gould (Rotenstein et al., 2008)), monoclonal PLP (Proteolipid protein, courtesy of D. Colman, (Sinoway et al., 1994), SMP (Schwann cell myelin protein), monoclonal 7B3/transitin, HNK-1 and 3A10 (DSHB, Developmental Studies Hybridoma Bank, UI), polyclonal Sox2 (Abcam), monoclonal Nf-M and CNPase (2', 3'-cyclic nucleotide 3'-phosphodiesterase) (Chemicon), polyclonal p75 (courtesy of L. Reichardt).
2.3 Wholemount immunofluorescence
Embryos were blocked overnight in blocking buffer (Phosphate Buffered Serum (PBS) with 1% Triton-X100, 10% Fetal Bovine Serum (FBS)), and then incubated with primary antibodies in PBS overnight at 4°C. The next day, embryos were extensively washed with PBS and incubated with secondary antibodies (anti-mouse or anti-rabbit-Alexa 594, Invitrogen, Molecular Probes). Because the nervous system of young embryos showed high levels of autofluorescence in the 488 wavelength we used 594 wavelength secondary antibodies whenever possible. The following day the embryos were washed extensively and photographed with either an A-1 AxioImager or a LUMAR by Zeiss. Since thin sections can only reach 35um thickness, we noticed this was not enough to visualize GFAP and FoxD3 immuno-fluorescence (Wotton et al., 2008), and used manual thick sections (1mm or less that one somite thickness) accordingly.
Supplementary Material
Acknowledgements
We have special thanks to Chris Plante and John Roesgen from the Long Beach Aquarium and Kiersten Darrow from Cabrillo Aquarium for providing the shark embryos. We thank Cindy Malone and Tatjana Sauka-Spengler for revising the manuscript, Jack Sechrist and Katy McCabe for useful discussions and Peter Rudy for technical assistance. To Bob Gould for providing several of the shark antibodies used in these paper. Partial support for M. Juarez participation on this project was provided by NIH GM 2 T34 GM008959 to ME Zavala. This work was partly supported by an NIH/NINDS AREA grant 1R15-NS060099-01 and NIH-MBRS SCORE-5S06GM048680-13 to MEdB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allinquant B, Staugaitis SM, D'Urso D, Colman DR. The ectopic expression of myelin basic protein isoforms in Shiverer oligodendrocytes: implications for myelinogenesis. J Cell Biol. 1991;113:393–403. doi: 10.1083/jcb.113.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ari C, Kalman M. Evolutionary changes of astroglia in Elasmobranchii comparing to amniotes: a study based on three immunohistochemical markers (GFAP, S-100, and glutamine synthetase) Brain Behav Evol. 2008a;71:305–24. doi: 10.1159/000129654. [DOI] [PubMed] [Google Scholar]
- Ari C, Kalman M. Glial architecture of the ghost shark (Callorhinchus milii, Holocephali, Chondrichthyes) as revealed by different immunohistochemical markers. J Exp Zoolog B Mol Dev Evol. 2008b;310:504–19. doi: 10.1002/jez.b.21223. [DOI] [PubMed] [Google Scholar]
- Bakay L, Lee JC. Ultrastructural changes in the edematous central nervous system. 3. Edema in shark brain. Arch Neurol. 1966;14:644–60. doi: 10.1001/archneur.1966.00470120076011. [DOI] [PubMed] [Google Scholar]
- Balfour FM. A preliminary account of the development of the elasmobranch fishes. Q. J. Microsc. Sci. 1874;4:323–364. [Google Scholar]
- Balfour FM. A treatise on comparative embryology. 2 v. Macmillan and Co.; London: 1880. [Google Scholar]
- Ballard WW, Mellinger Jean, Lechenault Henri. A Series of Normal Stages for Development of Scyliorhinus canicula, the Lesser Spotted Dogfish (Chondrichthyes: Scyliorhinidae) J Exp Zool. 1993;267:318–336. [Google Scholar]
- Brosamle C, Halpern ME. Characterization of myelination in the developing zebrafish. Glia. 2002;39:47–57. doi: 10.1002/glia.10088. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Cameron-Curry P, Dulac C, Le Dourain NM. A Monoclonal Antibody Defining a Carbohydrate Epitope Restricted to Glial Cells. Eur J Neurosci. 1991;3:126–139. doi: 10.1111/j.1460-9568.1991.tb00073.x. [DOI] [PubMed] [Google Scholar]
- Campbell K, Gotz M. Radial glia: multi-purpose cells for vertebrate brain development. Trends Neurosci. 2002;25:235–8. doi: 10.1016/s0166-2236(02)02156-2. [DOI] [PubMed] [Google Scholar]
- Cardone B, Roots BI. Comparative immunohistochemical study of glial filament proteins (glial fibrillary acidic protein and vimentin) in goldfish, octopus, and snail. Glia. 1990;3:180–92. doi: 10.1002/glia.440030305. [DOI] [PubMed] [Google Scholar]
- Chiba A. S-100 protein-immunoreactive structures in the brains of the elasmobranchs Scyliorhinus torazame and Mustelus manazo. Neurosci Lett. 2000;279:65–8. doi: 10.1016/s0304-3940(99)00949-0. [DOI] [PubMed] [Google Scholar]
- Cole NJ, Currie PD. Insights from sharks: evolutionary and developmental models of fin development. Dev Dyn. 2007;236:2421–31. doi: 10.1002/dvdy.21268. [DOI] [PubMed] [Google Scholar]
- Coolen A, Menuet D, Chassoux C, Compagnucci S, Henry L, Lévèque C, Da Silva F, Gavory S, Samain P, Wincker C, Thermes Y, D'Aubenton-Carafa G, Rodriguez-Moldes I, Naylor G, Depew M, Sourdaine P, Mazan S. The Dogfish Scyliorhinus canicula: A Reference in Jawed Vertebrates. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y.: 2009. [DOI] [PubMed] [Google Scholar]
- Csillag A, Kalman M. Evolutionary changes of astroglia in elasmobranchii comapring to ammniotes: a study based on three immunohistochemical markers (GFAP, S-100 and Glutamine synthetase) Brain Behav Evol. 2008;71:305–324. doi: 10.1159/000129654. [DOI] [PubMed] [Google Scholar]
- Dahl D, Crosby CJ, Sethi JS, Bignami A. Glial fibrillary acidic (GFA) protein in vertebrates: immunofluorescence and immunoblotting study with monoclonal and polyclonal antibodies. J Comp Neurol. 1985;239:75–88. doi: 10.1002/cne.902390107. [DOI] [PubMed] [Google Scholar]
- Derobert Y, Baratte B, Lepage M, Mazan S. Pax6 expression patterns in Lampetra fluviatilis and Scyliorhinus canicula embryos suggest highly conserved roles in the early regionalization of the vertebrate brain. Brain Res Bull. 2002;57:277–80. doi: 10.1016/s0361-9230(01)00695-5. [DOI] [PubMed] [Google Scholar]
- Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech. 2003;60:540–51. doi: 10.1002/jemt.10296. [DOI] [PubMed] [Google Scholar]
- Dottori M, Gross MK, Labosky P, Goulding M. The winged-helix transcription factor Foxd3 suppresses interneuron differentiation and promotes neural crest cell fate. Development. 2001;128:4127–38. doi: 10.1242/dev.128.21.4127. [DOI] [PubMed] [Google Scholar]
- Dulac C, Cameron-Curry P, Ziller C, Le Douarin NM. A surface protein expressed by avian myelinating and nonmyelinating Schwann cells but not by satellite or enteric glial cells. Neuron. 1988;1:211–20. doi: 10.1016/0896-6273(88)90141-9. [DOI] [PubMed] [Google Scholar]
- Dupin E, Baroffio A, Dulac C, Cameron-Curry P, Le Douarin NM. Schwann-cell differentiation in clonal cultures of the neural crest, as evidenced by the anti-Schwann cell myelin protein monoclonal antibody. Proc Natl Acad Sci U S A. 1990;87:1119–23. doi: 10.1073/pnas.87.3.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton JR, Antonellis A, Carney TJ, Rodrigues FS, Pavan WJ, Ward A, Kelsh RN. An evolutionarily conserved intronic region controls the spatiotemporal expression of the transcription factor Sox10. BMC Dev Biol. 2008;8:105. doi: 10.1186/1471-213X-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R, Cohn MJ. Analysis of EphA4 in the lesser spotted catshark identifies a primitive gnathostome expression pattern and reveals co-option during evolution of shark-specific morphology. Dev Genes Evol. 2004;214:466–72. doi: 10.1007/s00427-004-0426-0. [DOI] [PubMed] [Google Scholar]
- Freitas R, Zhang G, Albert JS, Evans DH, Cohn MJ. Developmental origin of shark electrosensory organs. Evol Dev. 2006;8:74–80. doi: 10.1111/j.1525-142X.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- Fu H, Qi Y, Tan M, Cai J, Takebayashi H, Nakafuku M, Richardson W, Qiu M. Dual origin of spinal oligodendrocyte progenitors and evidence for the cooperative role of Olig2 and Nkx2.2 in the control of oligodendrocyte differentiation. Development. 2002;129:681–93. doi: 10.1242/dev.129.3.681. [DOI] [PubMed] [Google Scholar]
- Gould RM, Fannon AM, Moorman SJ. Neural cells from dogfish embryos express the same subtype-specific antigens as mammalian neural cells in vivo and in vitro. Glia. 1995;15:401–18. doi: 10.1002/glia.440150405. [DOI] [PubMed] [Google Scholar]
- Gould RM, Spivack WD, Gilland E, Pant HC, Tseng D. Localization of Myelin Proteins in the Developing Shark Spinal Cord. Biol. Bull. 1992;183:358–359. doi: 10.1086/BBLv183n2p358. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Henion PD, Blyss GK, Luo R, An M, Maynard TM, Cole GJ, Weston JA. Avian transitin expression mirrors glial cell fate restrictions during neural crest development. Dev Dyn. 2000;218:150–9. doi: 10.1002/(SICI)1097-0177(200005)218:1<150::AID-DVDY13>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Jacobson M. Developmental neurobiology. Plenum Press; New York: 1991. p. ix.p. 776. [Google Scholar]
- Jacobson M, Rao MS. Developmental neurobiology. Kluwer Academic/Plenum; New York: 2005. p. xi.p. 424. [Google Scholar]
- Jessen KR, Mirsky R. The origin and development of glial cells in peripheral nerves. Nat Rev Neurosci. 2005;6:671–82. doi: 10.1038/nrn1746. [DOI] [PubMed] [Google Scholar]
- Kalman M. GFAP expression withdraws--a trend of glial evolution? Brain Res Bull. 2002;57:509–11. doi: 10.1016/s0361-9230(01)00713-4. [DOI] [PubMed] [Google Scholar]
- Kalman M, Gould RM. GFAP-immunopositive structures in spiny dogfish, Squalus acanthias, and little skate, Raia erinacea, brains: differences have evolutionary implications. Anat Embryol (Berl) 2001;204:59–80. doi: 10.1007/s004290100180. [DOI] [PubMed] [Google Scholar]
- Kaneko R, Sueoka N. Tissue-specific versus cell type-specific expression of the glial fibrillary acidic protein. Proc Natl Acad Sci U S A. 1993;90:4698–702. doi: 10.1073/pnas.90.10.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelsh RN, Dutton K, Medlin J, Eisen JS. Expression of zebrafish fkd6 in neural crest-derived glia. Mech Dev. 2000;93:161–4. doi: 10.1016/s0925-4773(00)00250-1. [DOI] [PubMed] [Google Scholar]
- Kettenmann H, Ransom BR. Neuroglia. Oxford University Press; New York: 2005. p. xix.p. 601. [Google Scholar]
- Kim H, Shin J, Kim S, Poling J, Park HC, Appel B. Notch-regulated oligodendrocyte specification from radial glia in the spinal cord of zebrafish embryos. Dev Dyn. 2008;237:2081–9. doi: 10.1002/dvdy.21620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kos R, Reedy MV, Johnson RL, Erickson CA. The winged-helix transcription factor FoxD3 is important for establishing the neural crest lineage and repressing melanogenesis in avian embryos. Development. 2001;128:1467–79. doi: 10.1242/dev.128.8.1467. [DOI] [PubMed] [Google Scholar]
- Kucenas S, Takada N, Park HC, Woodruff E, Broadie K, Appel B. CNS-derived glia ensheath peripheral nerves and mediate motor root development. Nat Neurosci. 2008;11:143–51. doi: 10.1038/nn2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani S, Horigome N. Developmental Morphology of Branchiomeric Nerves in a Cat Shark, Scyliorhinus torazame, with Special Reference to Rhombomeres, Cephalic Mesoderm, and Distribution Patterns of Cephalic Crest Cells. ZOOLOGICAL SCIENCE. 2000;17:893–909. [Google Scholar]
- Lazzari M, Franceschini V. Glial fibrillary acidic protein and vimentin immunoreactivity of astroglial cells in the central nervous system of the African lungfish, Protopterus annectens (Dipnoi: Lepidosirenidae) J Morphol. 2004;262:741–9. doi: 10.1002/jmor.10274. [DOI] [PubMed] [Google Scholar]
- Levitt P, Cooper ML, Rakic P. Early divergence and changing proportions of neuronal and glial precursor cells in the primate cerebral ventricular zone. Dev Biol. 1983;96:472–84. doi: 10.1016/0012-1606(83)90184-7. [DOI] [PubMed] [Google Scholar]
- Lister JA, Cooper C, Nguyen K, Modrell M, Grant K, Raible DW. Zebrafish Foxd3 is required for development of a subset of neural crest derivatives. Dev Biol. 2006;290:92–104. doi: 10.1016/j.ydbio.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Lunn ER, Scourfield J, Keynes RJ, Stern CD. The neural tube origin of ventral root sheath cells in the chick embryo. Development. 1987;101:247–54. doi: 10.1242/dev.101.2.247. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–64. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Martenson RE. Myelin--biology and chemistry. CRC Press; Boca Raton, FL: 1992. p. 958. [Google Scholar]
- Morell P. Myelin. Plenum Press; New York: 1984. p. xx.p. 545. [Google Scholar]
- Nakao T, Ishizawa A. Development of the spinal nerves in the lamprey: III. Spinal ganglia and dorsal roots in 26-day (13 mm) larvae. J Comp Neurol. 1987;256:369–85. doi: 10.1002/cne.902560306. [DOI] [PubMed] [Google Scholar]
- Ndubaku U, de Bellard ME. Glial cells: old cells with new twists. Acta Histochem. 2008;110:182–95. doi: 10.1016/j.acthis.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill P, McCole RB, Baker CV. A molecular analysis of neurogenic placode and cranial sensory ganglion development in the shark, Scyliorhinus canicula. Dev Biol. 2007;304:156–81. doi: 10.1016/j.ydbio.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onteniente B, Kimura H, Maeda T. Comparative study of the glial fibrillary acidic protein in vertebrates by PAP immunohistochemistry. J Comp Neurol. 1983;215:427–36. doi: 10.1002/cne.902150407. [DOI] [PubMed] [Google Scholar]
- Park HC, Shin J, Appel B. Spatial and temporal regulation of ventral spinal cord precursor specification by Hedgehog signaling. Development. 2004;131:5959–69. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- Potzner MR, Griffel C, Lutjen-Drecoll E, Bosl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–26. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J Comp Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–6. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Riuzzi F, Sorci G, Donato R. S100B stimulates myoblast proliferation and inhibits myoblast differentiation by independently stimulating ERK1/2 and inhibiting p38 MAPK. J Cell Physiol. 2006;207:461–70. doi: 10.1002/jcp.20580. [DOI] [PubMed] [Google Scholar]
- Romero-Aleman Mdel M, Monzon-Mayor M, Yanes C, Arbelo-Galvan JF, Lang D, Renau-Piqueras J, Negrin-Martinez C. S100 immunoreactive glial cells in the forebrain and midbrain of the lizard Gallotia galloti during ontogeny. J Neurobiol. 2003;57:54–66. doi: 10.1002/neu.10258. [DOI] [PubMed] [Google Scholar]
- Rotenstein L, Herath K, Gould RM, de Bellard ME. Characterization of the shark myelin Po protein. Brain Behav Evol. 2008;72:48–58. doi: 10.1159/000145717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovainen CM. Neurobiology of lampreys. Physiol Rev. 1979;59:1007–77. doi: 10.1152/physrev.1979.59.4.1007. [DOI] [PubMed] [Google Scholar]
- Sanosaka T, Namihira M, Asano H, Kohyama J, Aisaki K, Igarashi K, Kanno J, Nakashima K. Identification of genes that restrict astrocyte differentiation of midgestational neural precursor cells. Neuroscience. 2008;155:780–8. doi: 10.1016/j.neuroscience.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Baratte B, Shi L, Mazan S. Structure and expression of an Otx5- related gene in the dogfish Scyliorhinus canicula: evidence for a conserved role of Otx5 and Crxgenes in the specification of photoreceptors. Dev Genes Evol. 2001;211:533–44. doi: 10.1007/s00427-001-0191-2. [DOI] [PubMed] [Google Scholar]
- Schmechel DE, Rakic P. Arrested proliferation of radial glial cells during midgestation in rhesus monkey. Nature. 1979;277:303–5. doi: 10.1038/277303a0. [DOI] [PubMed] [Google Scholar]
- Schweigreiter R, Roots BI, Bandtlow CE, Gould RM. Understanding myelination through studying its evolution. Int Rev Neurobiol. 2006;73:219–73. doi: 10.1016/S0074-7742(06)73007-0. [DOI] [PubMed] [Google Scholar]
- Sinoway MP, Kitagawa K, Fidler L, Gould RM, Colman DR. Tissue lipoproteins revisited: new proteolipid protein gene family members in elasmobranchs. Neurochem Res. 1994;19:1047–54. doi: 10.1007/BF00968715. [DOI] [PubMed] [Google Scholar]
- Snipes GJ, Suter U, Shooter EM. The genetics of myelin. Curr Opin Neurobiol. 1993;3:694–702. doi: 10.1016/0959-4388(93)90140-t. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Melton KR, Manzanares M. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int J Dev Biol. 2003;47:541–53. [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S. Ancient noncoding elements conserved in the human genome. Science. 2006;314:1892. doi: 10.1126/science.1130708. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S. Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 2007;5:e101. doi: 10.1371/journal.pbio.0050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waehneldt TV. Phylogeny of myelin proteins. Ann N Y Acad Sci. 1990;605:15–28. doi: 10.1111/j.1749-6632.1990.tb42377.x. [DOI] [PubMed] [Google Scholar]
- Waehneldt TV, Matthieu JM, Stoklas S. Immunological evidence for the presence of myelin-related integral proteins in the CNS of hagfish and lamprey. Neurochem Res. 1987;12:869–73. doi: 10.1007/BF00966308. [DOI] [PubMed] [Google Scholar]
- Wang SZ, Dulin J, Wu H, Hurlock E, Lee SE, Jansson K, Lu QR. An oligodendrocyte-specific zinc-finger transcription regulator cooperates with Olig2 to promote oligodendrocyte differentiation. Development. 2006;133:3389–98. doi: 10.1242/dev.02522. [DOI] [PubMed] [Google Scholar]
- Wicht H, Derouiche A, Korf HW. An immunocytochemical investigation of glial morphology in the Pacific hagfish: radial and astrocyte-like glia have the same phylogenetic age. J Neurocytol. 1994;23:565–76. doi: 10.1007/BF01262057. [DOI] [PubMed] [Google Scholar]
- Woodruff RH, Tekki-Kessaris N, Stiles CD, Rowitch DH, Richardson WD. Oligodendrocyte development in the spinal cord and telencephalon: common themes and new perspectives. Int J Dev Neurosci. 2001;19:379–85. doi: 10.1016/s0736-5748(00)00083-6. [DOI] [PubMed] [Google Scholar]
- Wotton KR, Mazet F, Shimeld SM. Expression of FoxC, FoxF, FoxL1, and FoxQ1 genes in the dogfish Scyliorhinus canicula defines ancient and derived roles for Fox genes in vertebrate development. Dev Dyn. 2008;237:1590–603. doi: 10.1002/dvdy.21553. [DOI] [PubMed] [Google Scholar]
- Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–90. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Colman DR. Parallel evolution and coexpression of the proteolipid proteins and protein zero in vertebrate myelin. Neuron. 1996;16:1115–26. doi: 10.1016/s0896-6273(00)80138-5. [DOI] [PubMed] [Google Scholar]
- Zalc B. The acquisition of myelin: a success story. Novartis Found Symp. 2006;276:15–21. doi: 10.1002/9780470032244.ch3. discussion 21-5, 54-7, 275-81. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.