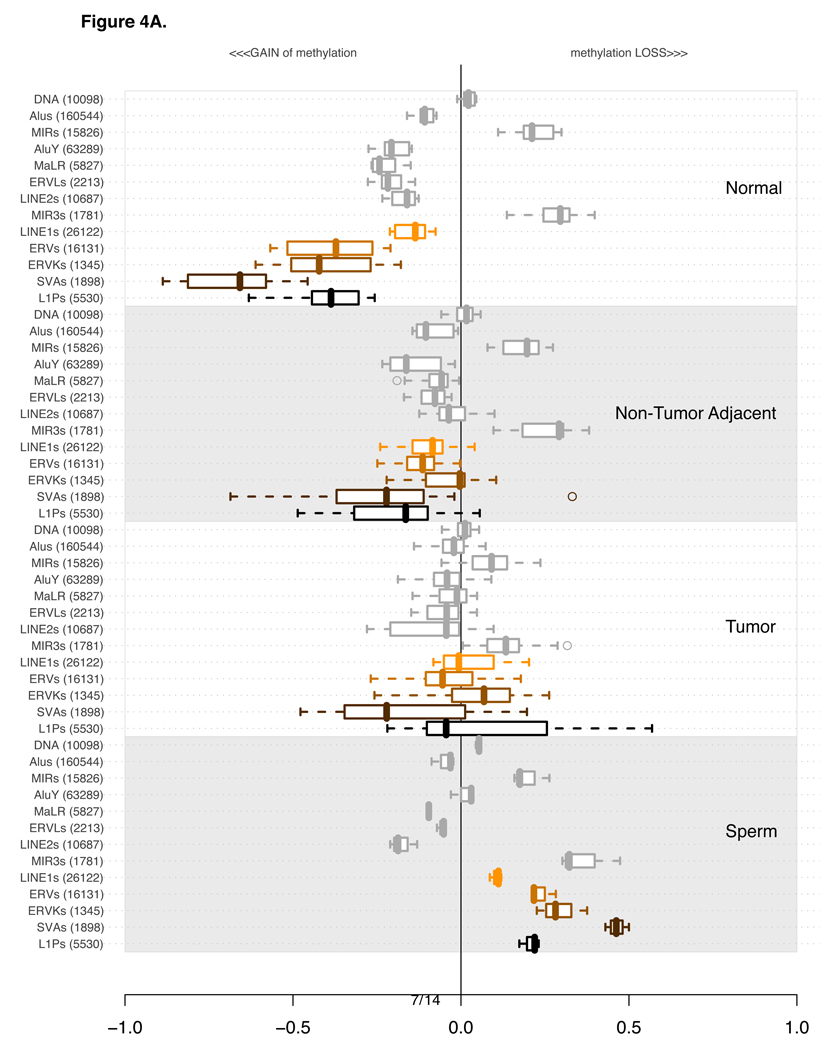
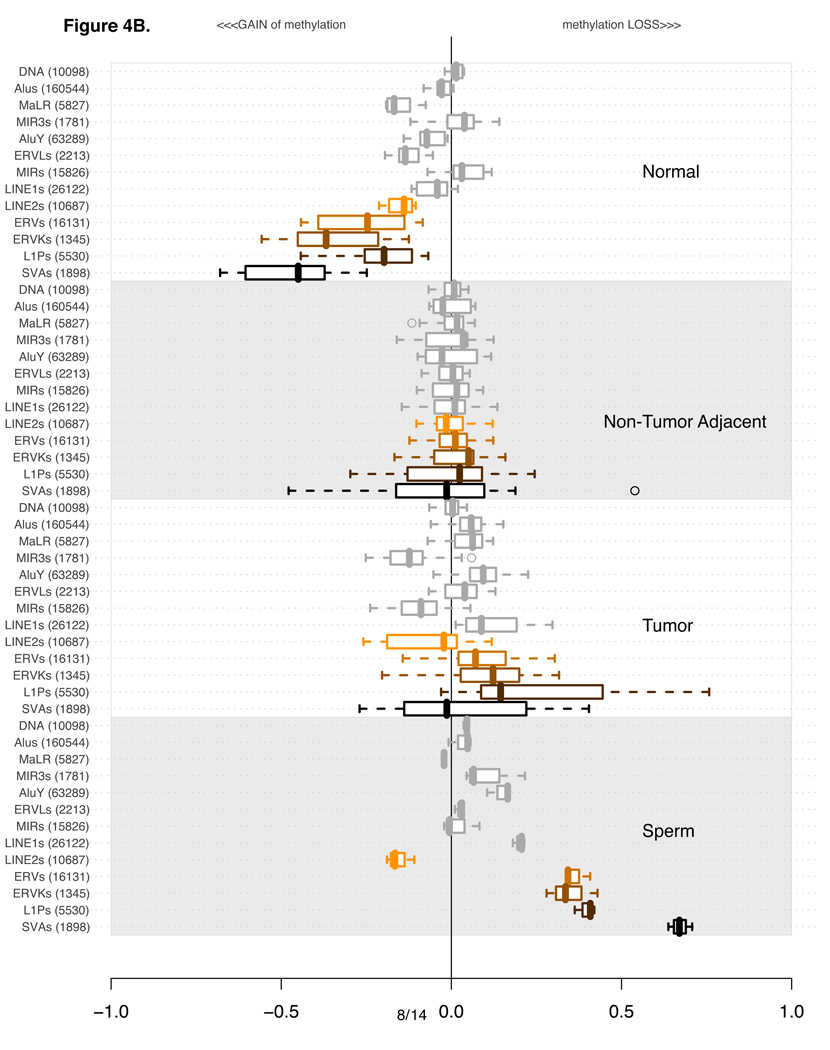
Figure 4.
The four sections of the plot indicate 4 distinct classes of tissue types used for methylation profiling: Normals (10 experiments), non-tumor adjacent (17 experiments), Tumors (33 Experiments) and Sperm (3 replicate experiments). Each of the four sections contains the methylation levels of the same 13 categories of repetitive elements.
Per category, the values are summarized using a box-and-whisker plot. A line within each box indicates the median value. Box boundaries are drawn based on 1st and 3rd quartiles. The dashed lines extending from the box indicate the extreme values of the distribution. Outliers, if any, are indicated by a circle.
The classes and families of repetitive elements are indicated on the left of the box-and-whisker segment. The number in parenthesis next to the category description indicates the number of probes corresponding to the number of repetitive elements uniquely probed in the genome.
The order of categories is constant in all four of the subsections. It was established based on the extent of variation in the plotted distributions using Shannon entropy information content metric. Only Normal and Tumor experiments were used to calculate the Shannon’s Information metric. For a more detailed explanation see Supplementary Section 3.
A) Distribution of average methylation levels per category. In each of the 4 subsections of the plot the pertinent experiments contributed an average methylation level for all probes in proximity of a specific class of repetitive element.
B) As in part A, except this time every experiment is normalized using an average of all tumor-adjacent experiments.