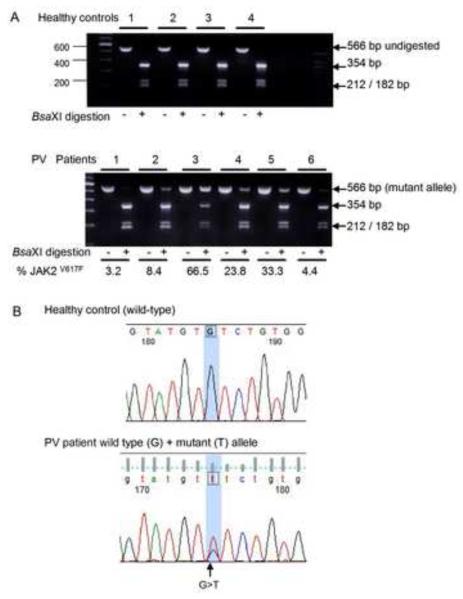
Figure 1. Detection of JAK2V617F mutation in primary erythroid precursors.
Total RNA isolated from primary erythroblasts of healthy controls and PV patients was reverse transcribed followed by PCR amplification for JAK2 yielding a 566 bp product. (A) Detection of JAK2V617F by restriction enzyme BsaXI digestion. The wild-type allele is digested into 354, 212, and 182 bp fragments whereas the mutant allele, detected only in PV patients, remains undigested. The allele-burden of JAK2V617F in PV patients was quantified by densitometry and expressed as percentage. (B) Sequence traces illustrating wild-type sequence (a healthy control) and confirmation of mixed G>T sequences in a representative patient with PV. The presence of the mutant JAK2V617F allele was confirmed by direct sequencing in all patient samples.