Figure 1.
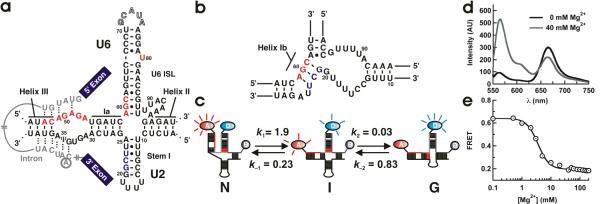
(a) Secondary structure model of the spliceosomal snRNAs U2/U6 from S. cerevisiae with an intron bound.19 Highly conserved residues in U6 (ACAGAGA loop, AGC triad and U80) are highlighted in red. U2 residues involved in helix IB formation are highlighted in grey. The U6 ISL pentaloop (outlined residues G71–A75) was deleted to facilitate construct labeling. (b) Proposed structure of helix IB. (c) Proposed folding reaction pathway for the U2/U6 complex (rates measured at 40 mM Mg2+, units are s−1). At least three distinct conformations are observed for the U2/U6 snRNA complex: a high FRET conformation that resembles the 4-helix structure (N), a low FRET conformation that resembles the 3-helix structure with helix IB (G), and a previously unobserved folding intermediate (I). (d) Mg2+-induced conformational change in the U2/U6 spliceosomal complex. Fluorescence emission spectra of the fluorophore labeled U2/U6 complex, in the absence (black) and presence (grey) of Mg2+ ions. The donor fluorophore (Cy3) emits at 565 nm and the acceptor (Cy5) at 665 nm. Mg2+ ions decrease the acceptor emission and increase the donor emission, revealing the presence of a conformational change. (e) Calculated FRET ratio as a function of [Mg2+]. The line is a fit to a modified Hill equation (KMg = 3.0 ± 0.2 mM and n = 2.2 ± 0.2).