Figure 4. Toeprinting and primer extension analyses.
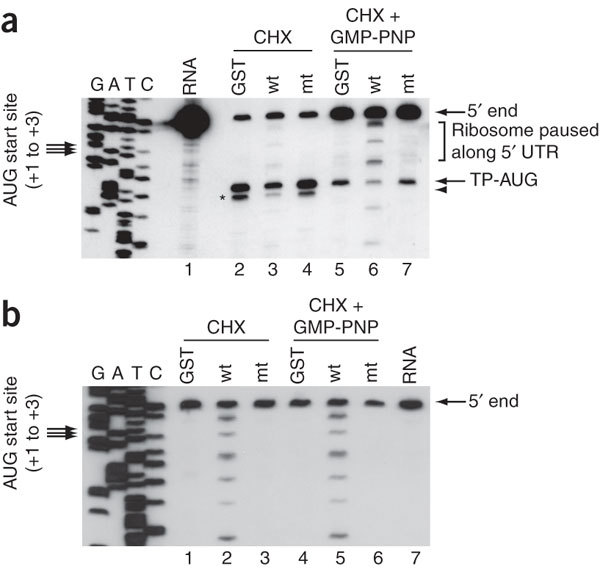
(a) Toeprinting. Samples shown in lanes 2–4 and 5–7 were incubated with CHX and a mixture of CHX and GMP-PNP, respectively, and toeprinting analysis was performed in the presence of GST (lanes 2 and 5), nsp1 (lanes 3 and 6) or nsp1-mt (lanes 4 and 7). Lane 1, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer; 5′ end, the primer extension product that was extended to the 5′ end of rLuc RNA; UTR, untranslated region; TP-AUG, a correctly positioned toeprint. Arrowhead and asterisk represent a signal generated by the joining of the 60S subunits to the 40S complex positioned at the AUG codon. A dideoxynucleotide sequence of the rLuc gene generated with the same primer was run in parallel (left four rows).Translation initiation codon is shown at the left side of the gel. (b) Primer extension analysis. RRL was incubated with GST (lanes 1 and 4), nsp1 (lanes 2 and 5), or nsp1-mt (lanes 3 and 6) in the presence of CHX (lanes 1–3) or a mixture of CHX and GMP-PNP (lanes 4–6) for 5 min at 30 °C. Then rLuc RNA was added to each sample, and the samples were incubated for another 10 min at 30 °C. The RNAs were extracted and subjected to primer extension analysis. Lane 7, the primer extension products using the rLuc RNA that was hybridized with a 5′-end labeled primer.
