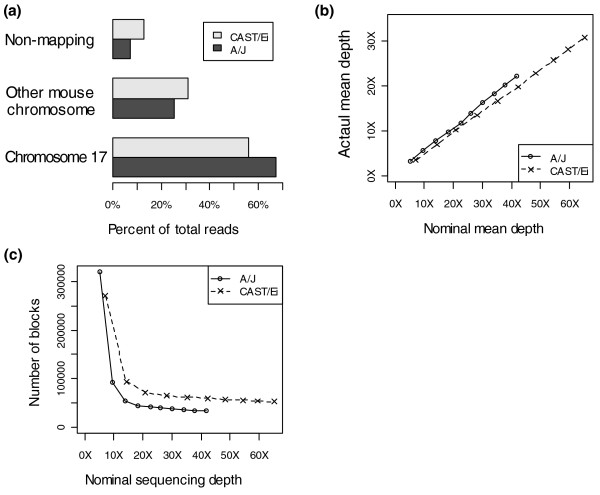
Figure 1.
Mapping of short read sequence to the mouse genome. The MAQ algorithm was used to map the short read sequences to the NCBI 37 mouse genome assembly. (a) The percentage of reads that map to chromosome 17, other mouse chromosomes, or not at all to the C57BL/6J reference assembly. (b) The actual average sequence depth over chromosome 17 after duplicate sequence reads have been removed, plotted against the nominal depth if all reads were unique and mapped to chromosome 17. (c) The number of contiguous blocks of sequence, defined as a stretch of sequence where all bases have non-zero sequencing depth over them, plotted against nominal depth (see above).