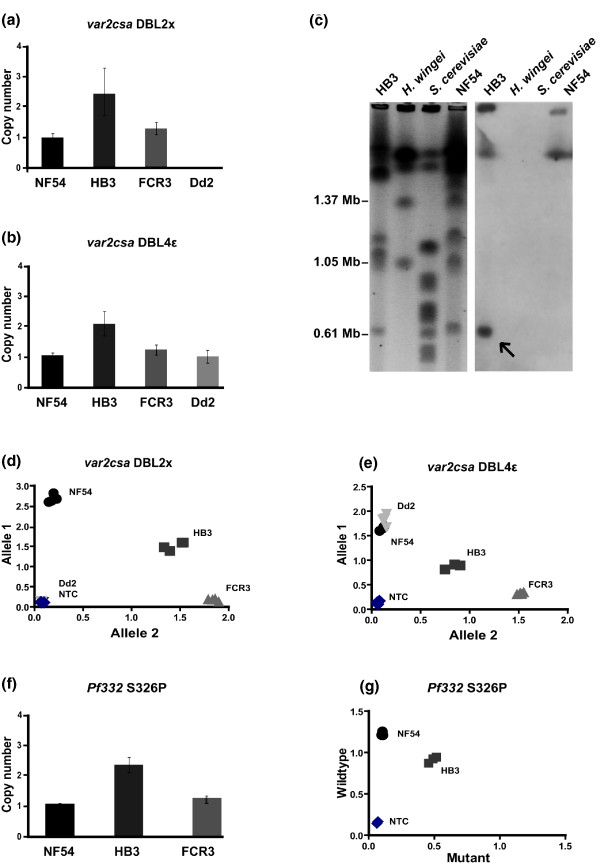
Figure 3.
Copy numbers of var2csa and Pf332 alleles and allelic discrimination. (a, b, f) var2csa and Pf332 gene copy numbers in various parasite strains are shown relative to the NF54 strain, with error bars representing the confidence interval (CI 95%). Two gene copies were identified in all cases for the HB3 parasite, suggesting that the three additional DBL4ε sequences are due to the partial assembly of this fully sequenced genome. (c) PFGE followed by Southern blotting revealed the second var2csa copy to be located on chromosome 1 in HB3. An ethidium bromide stained PFGE gel is shown on the left with separated chromosomes from HB3, NF54 and the standards Hansenula wingei and Saccharomyces cerevisiae; selected chromosome sizes are indicated in megabase-pairs (Mb). The Southern blot shown on the right revealed the var2csa-specific DNA probe to hybridize to chromosome 1 in HB3 (indicated with an arrow). (d, e) Discrimination of var2csa alleles in gDNA from the indicated parasites showed single allele frequency in NF54, Dd2 (Allele 1) and FCR3 (Allele 2), and double alleles in HB3. (g) The same analysis on the S326P mutation in Pf332 revealed only the wild-type version in NF54, whereas HB3, with its dual copies, harbors both the wild-type and mutant versions.