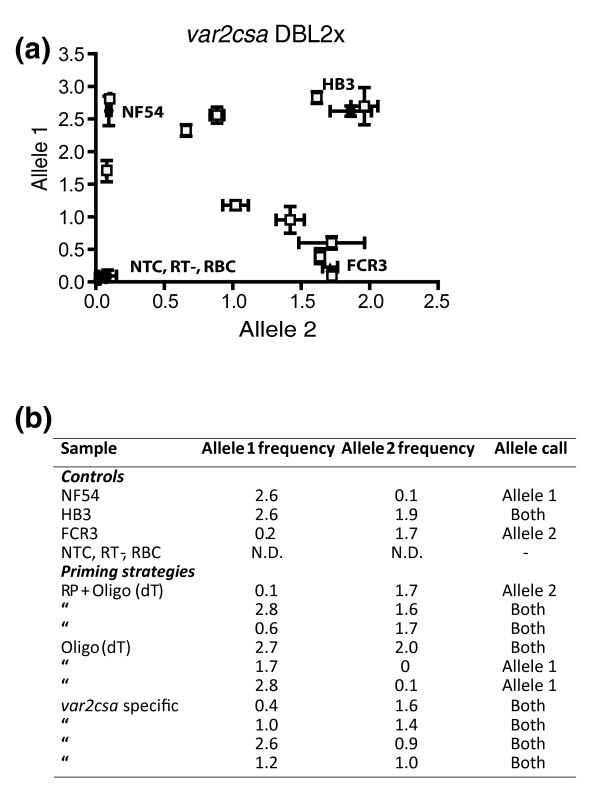
Figure 5.
var2csa allele transcriptional activity in individual HB3CSA parasites. Single cell transcription from 11 individual HB3 parasites repeatedly selected for CSA-binding phenotype. Parasites at the mature trophozoite stage (24 ± 4 h p.i.) were subjected to three different priming strategies during the reverse transcription (random primers and oligo(dT), oligo(dT) only and var2csa-specific primers). (a) Allele frequencies of alleles 1 and 2 for the positive gDNA controls NF54 (filled circles), FCR3 (filled triangles) and HB3 (filled squares) as well as for the 11 cDNA samples (empty square). Negative controls (filled diamonds) with RNA reverse transcribed without addition of reverse transcriptase (RT-), exchange of template for ddH2O (NTC) and amplifications from uninfected red blood cells (RBCs) were included in all experiments to prevent signals from gDNA, contaminations or unspecific amplifications influencing the interpretation of the results. All data-points represent means of triplicates with standard deviations for each allele expressed as bi-directional error bars. (b) Mean allele frequencies and predicted allele calls for all samples and priming strategies described above. N.D., not detected.