Abstract
Peptidase inhibitor 3 (PI3, elafin) is a protease inhibitor produced locally in the lung, where it plays a central role in controlling excessive activity of neutrophil elastase. Our previous study revealed that PI3 gene expression is down-regulated during the acute stage of acute respiratory distress syndrome (ARDS). We conducted a case-control study to investigate whether genetic variants in PI3 gene are associated with ARDS development. Based on resequencing data from 29 unrelated white subjects, three tagging single-nucleotide polymorphisms were selected and genotyped in a prospective cohort consisting of 449 white patients with ARDS (cases) and 1,031 critically ill patients (at-risk control subjects). We found that the variant allele of rs2664581 (T34P) was significantly associated with increased ARDS risk (odds ratio [OR], 1.35; 95% confidence interval [CI], 1.09–1.67; P = 0.006; false discovery rate adjusted P = 0.018). Moreover, this association was stronger among subjects with extrapulmonary injury. The common haplotype Hap2 (TTC), containing the variant allele of rs2664581, was also identified as a risk haplotype for ARDS (OR, 1.31; 95% CI, 1.05–1.64; P = 0.015). Furthermore, the rs2664581 polymorphism was associated with circulating PI3 levels in multivariate analyses. Patients with ARDS homozygous for the wild-type A allele of rs2664581 showed significant lower PI3 plasma level (P = 0.019) at ARDS onset as compared with those homozygous or heterozygous for the variant C allele. Our data suggest that polymorphisms in PI3 gene are significantly associated with ARDS risk and with circulating PI3 levels.
Keywords: acute respiratory distress syndrome, elafin, genetic susceptibility, haplotypes, tagging SNP
CLINICAL RELEVANCE
The present study demonstrates a significant association between peptidase inhibitor 3 (PI3) polymorphisms and risk for acute respiratory distress syndrome (ARDS). The single-nucleotide polymorphisms in PI3 gene may be important in controlling the PI3 plasma levels and the capability to protect from neutrophil elastase–induced lung injury. Our findings may have significant implications for risk assessment and treatment of patients with ARDS.
Acute respiratory distress syndrome (ARDS) is characterized by inflammation of lung parenchyma induced by either direct pulmonary insults or secondary responses to extrapulmonary injuries. ARDS starts with damage to the integrity of the alveolar–capillary barrier resulting in leaking plasma and inflammatory cells into the interstitium and alveolar space, and eventually leading to impaired gas exchange (1, 2). Previous studies demonstrated that neutrophils and their derived proteinases play crucial roles in initiating and propagating pulmonary damage (3, 4). Human neutrophil elastase (HNE) is a serine protease used by neutrophils to degrade extracellular matrix (ECM) components and facilitate its migration through the tissues to the inflammatory site. Under physiologic conditions, HNE participates in host defense against microbial pathogens and is tightly regulated by endogenous protease inhibitors. At inflammatory sites, however, HNE appears to remain active and cause tissue damage, due to an imbalance between elevated HNE level and that of the endogenous protease inhibitors (4).
Peptidase inhibitor 3 (PI3) (also known as elastase specific inhibitor, skin-derived antileukoprotease, or elafin) is a low-molecular-weight proteinase inhibitor that is synthesized and secreted at the site of neutrophil infiltration. The expression of PI3 gene can be induced by proinflammatory cytokines (IL-1β, TNF-α), bacterial lipopolysaccharides, and elastase (5–7), and there is constitutive expression in airways and other mucosal sites where the inflammatory stimuli persist (8). In contrast to other protease inhibitors, such as secretory leukocyte proteinase inhibitor and α1-antitrypsin, PI3 has a narrower spectrum of inhibition specifically toward HNE (9). Moreover, PI3 protein has a unique molecular structure comprising two domains, including a carboxy-terminal whey acidic protein (WAP) domain containing the antiprotease active site and an amino-terminal domain (38 residues) containing several transglutaminase substrate motifs characterized by hexapeptide repeats with a consensus sequence GQDPVK (10, 11). The transglutaminase substrate motifs enable covalent cross-linking of PI3 protein to ECM proteins, where PI3 can still exert its inhibitory function against HNE (12). These features make PI3 maximally effective as a tissue-bound inhibitor in the pulmonary interstitial environment by providing efficient protection against deregulated HNE activity. Previous studies demonstrate that human PI3 has significant protective effects against human NE-induced acute lung injury in several animal models (9). In addition to its role as protease inhibitor, PI3 has also been demonstrated to have many biological functions including antimicrobial, anti-inflammatory, and antiviral activities and immunomodulatory functions (9, 13).
Recently, our genome-wide expression analysis revealed that the expression of PI3 gene in peripheral blood is down-regulated during the initial stage of ARDS (Day 1 to 3 of diagnosis) as compared with the recovery stage (14). Consistent with the microarray findings, enzyme-linked immunosorbent assays revealed a rapid decrease of plasma PI3 levels during the initial stage of ARDS compared with PI3 levels within a 3-day period before ARDS diagnosis. These results suggest that PI3 might play a protective role in the early development of ARDS. Therefore, we hypothesized that polymorphisms in PI3 gene may be associated with the risk of developing ARDS. In the present study, we identified the common polymorphisms in PI3 gene through sequencing the entire genomic region containing PI3 gene in 29 unrelated white subjects. Next, we evaluated the association between PI3 polymorphisms and ARDS using a tagging single-nucleotide polymorphism (tSNP) approach in a large case-control study to test our hypothesis. Since previous evidence suggested that the genetic association with ARDS might be modified by the origin of lung injury (15, 16), we conducted stratified analyses among patients with different types of lung injury (pulmonary versus extrapulmonary lung injury). Finally, we investigated the relationship between PI3 polymorphisms and PI3 plasma levels at the onset of ARDS.
MATERIALS AND METHODS
Study Population
All subjects were recruited from patients admitted to the intensive care units (ICU) of the Massachusetts General Hospital (Boston, MA) between September 1999 and November 2006. Details of the recruitment have been described previously (17). Briefly, patients with pre-depositing conditions for ARDS development and without any of the exclusion criteria (see Table E1 in the online supplement) were enrolled and followed daily for the development of ARDS, as defined by the American-European Consensus Committee (AECC) criteria for ARDS (18). At-risk patients who did not meet the criteria for ARDS during the ICU hospitalization were classified as control subjects. The flowchart of study design is illustrated in Figure E1. Baseline characteristics were recorded on ICU admission, and Acute Physiology Age and Chronic Health Evaluation (APACHE) III scores were calculated based on the data collected within the first 24 hours after ICU admission. We defined pneumonia, pulmonary contusion, and aspiration as pulmonary risk factor for ARDS (pulmonary injury), whereas we defined sepsis and/or bacteremia originated from extrapulmonary sources, nonpulmonary trauma, and multiple transfusion as extrapulmonary risk factors (extrapulmonary injury). All patients with ARDS were followed for clinical outcomes of all-cause mortalities for 60 days after ARDS diagnosis. To reduce possible interference from population stratification, all analyses were restricted to the non-Hispanic white subjects, who account for 90.7% of the studied population. This study was approved by the Human Subjects Committees of the Massachusetts General Hospital and the Harvard School of Public Health. An informed consent was obtained from each enrolled patient or an appropriate proxy.
Identification of Genetic Variations at PI3 Locus
The gene PI3 located in chromosome 20 (20q12-q13) is approximately 2.3 kb in length and is described as containing three exons and two introns (19). We resequenced 2 kb of the 5′and 3′untranslated regions and all exons and introns using DNA samples from 29 nonrelated, healthy white subjects recruited as control subjects from a lung cancer susceptibility study. These participants were healthy friends or spouses of lung cancer cases. This sample size provides a greater than 99% detection rate to identify polymorphisms with minor allele frequency (MAF) greater than 5% (20). Bidirectional sequence analyses were conducted at the Harvard-Partners Center for Genetics and Genomics, Boston. GenBank accession no. AL049767.12 sequence was used as the reference sequence for defining the sequence variants. Linkage disequilibrium (LD) blocks of the PI3 gene were determined by SNPs with MAF greater than or equal to 10%. Genotypes of one SNP (PI3–1) could not be determined by sequencing in over 50% of the subjects, so it was also excluded from the LD block determination. Haploview (v3.32) software was used to determine the LD structure as described previously (21, 22). To efficiently tag the common variation across the gene, we selected tSNPs using the r2-based Tagger program (23) implemented in Haploview software, with the aggressive mode and the Tagger thresholds of r2 ≥ 0.8 and LOD score greater than 3.
Genotyping
Genomic DNA was extracted from whole blood sample using Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) or the AutoPure LS workstation using the Autopure reagents (Qiagen, Valencia, CA). SNPs rs19836491, rs6032040, and rs2664581 were genotyped using TaqMan SNP Genotyping Assay (Applied Biosystems, Foster City, CA). Primers and probes were ordered from Applied Biosystems (TaqMan Assay ID: C__25619045_10, C__29073396_10, and C__11656453_1_, respectively), and genotyping was performed using the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Laboratory personnel were blinded to case-control status. To ensure the quality of genotyping data, 10% of randomly selected samples were interspersed in the plates as replicates and all genotyping results were reviewed by two investigators independently. Based on comparison of replicate genotypes, the genotyping error rate was less than 0.05% and the overall genotyping success rate was above 98%. Samples not yielding the genotypes of all SNPs were excluded from analysis.
ELISA Analysis of Plasma PI3 Level
In addition to blood samples for DNA extraction, plasma samples were also collected for long-term storage, which were used in the present study. Based on the original sample protocol, plasma was collected from each enrolled patients within 48 hours of ICU admission. If an enrolled patient developed ARDS, an additional sample was collected within the first 48 hours of ARDS diagnosis. However, given the critical condition of ICU patients, heavy and intense workload for ICU research staff, difficulties of obtaining consent from patients' proxies in time, and strict limitation of total blood drawn from each subject set up by the IRB committee, only a fraction of patients with ARDS provided multiple plasma samples. In the current study, we used all available ARDS samples (n = 54) and 60 randomly selected plasma samples from the control subjects, including nine nonwhite subjects (six cases and three control subjects). For the plasma protein analysis portion of the study, considering the relatively small number of plasma samples, we included nonwhite samples in the study with the adjustment for ethnicity in the analyses. Plasma samples were stored at −80°C until analysis. Plasma PI3 levels were quantified in duplicate using Human pre-ELAFIN/SKALP (Cat. No. HK318) ELISA Test Kit (Cell Sciences, Canton, MA), according to the manufacturer's protocol. The coefficients of variations of duplicate samples were under 13%, which were below the manufacturer's recommended 15% standard.
Statistical Analysis
All statistical analyses were performed using the SAS statistical software package (version 9.1; SAS, Cary, NC). A P value < 0.05 was considered to be statistically significant. The demographic variables between patients with ARDS and control subjects were tested by Fisher's exact test for categorical variables and by Student's t test for continuous variables. We used SAS/Genetics PROC ALLELE test to calculate the allele frequencies, to test the deviation from Hardy-Weinberg equilibrium (HWE), and to estimate the pairwise D′ and r2 values for LD. Haplotype frequencies and individual haplotypes were estimated based on unphased genotype data using the expectation maximization algorithm as implemented in SAS/Genetics. The associations between PI3 haplotypes and the risk of ARDS were analyzed using the expectation-substitution approach as implemented in the SAS macro HAPPY (24, 25). This approach treats subject-specific expected haplotype indicators, calculated by an additive model, as observed covariates for regression models. Haplotypes with a frequency greater than 5% in the total population were considered as “common” and the most common haplotypes was used as the reference in the logistic regression models.
We used multivariate logistic regression to estimate the genotype- and haplotype-specific odds ratio (OR) and 95% confidence interval (CI) for ARDS susceptibility. The genotype associations were analyzed in both additive and dominant models. To evaluate the associations of individual genotypes and haplotypes with the survivals in patients with ARDS, we use the Cox proportional hazard model to estimate the hazard ratio (HR) and 95% CI. Covariates were chosen based on the potential risk for ARDS development and mortality, including age, sex, APACHE III score, and risk factors for ARDS, comorbidities, and alcohol abuse. Global test for the association between haplotypes and ARDS risk and survival were performed using the likelihood ratio test (LRT), comparing the models with the haplotypes to the models without. For statistically significant associations, adjusted P values were calculated to correct for multiple comparisons, using the False Discovery Rate (FDR) procedure described by Benjamini and colleagues (26). The distribution of plasma PI3 levels was evaluated by Kolmogorov-Smirnov and Anderson-Darling methods. Since plasma profiles of PI3 had skewed distributions, the natural log transformed data were used in the analyses. Plasma PI3 levels between ARDS cases and control subjects were compared using two sample t test and general linear model (GLM).
RESULTS
Characteristics of Study Population
During the study period, a total of 1,651 patients were enrolled into the prospective cohort. The final analyses were conducted in 1,480 non-Hispanic white subjects with complete clinical and genotyping data, including 449 ARDS cases and 1,031 at-risk control subjects. The distribution of the baseline characteristics of the study population and the comparisons of co-morbidities and clinical risk factors for ARDS between groups are shown in Table 1. Subjects who developed ARDS were younger and have higher APACHE III scores. Proportion of subjects on ventilation at ICU admission or with septic shock, pneumonia, pulmonary injury, liver cirrhosis/failure, and history of alcohol abuse was higher in the ARDS group, whereas sepsis without septic shock and diabetes were more common in the control subjects. No differences in sex distribution were observed between patients with ARDS and control subjects. When the patients with ARDS with plasma samples were compared with those without plasma samples, there were no significant differences with regard to age, sex, or other relevant baseline characteristics (P > 0.05 for all comparisons). However, the studied ARDS cases had significantly lower proportions of white subjects as well as higher APACHE III score on ICU admission (Table E6). Fifty-nine critically ill patients with ARDS risk factors randomly selected as control subjects had baseline characteristics that were not significantly different from the unselected control subjects, except that selected control subjects had lower proportions of white subjects (P > 0.05 for all comparisons) (Table E7). Since the paired plasma from one control failed to generate reliable plasma profiles, we excluded this subject resulting in 59 control subjects in the analyses. Baseline characteristics between ARDS cases and control subjects with available plasma samples are shown in Table E2. A difference in corticosteroid treatment was detected between ARDS and control with available plasma samples.
TABLE 1.
CHARACTERISTICS OF THE STUDY POPULATION
| Characteristic | Control (n = 1,031) | ARDS (n = 449) | P Value |
|---|---|---|---|
| Age, yr (mean ± SD) | 62.78 ± 17 | 59.49 ± 18 | <0.001 |
| Male, n (%) | 627 (60.8%) | 269 (59.9%) | 0.744 |
| APACHE III score, mean ± SD* | 66.46 ± 23 | 77.40 ± 24 | <0.001 |
| On ventilation at ICU admission, n (%) | 675 (65.5%) | 392 (87.3%) | <0.001 |
| Risk factors, n (%) | |||
| Sepsis | 383 (37.1%) | 117 (26.1%) | <0.001 |
| Septic shock | 448 (43.4%) | 267 (59.5%) | <0.001 |
| Pneumonia | 443 (43.0%) | 302 (67.3%) | <0.001 |
| Aspiration | 86 (8.3%) | 45 (10.0%) | 0.295 |
| Multiple transfusion | 118 (11.4%) | 47 (10.5%) | 0.583 |
| Trauma | 80 (7.8%) | 32 (7.1%) | 0.672 |
| Pulmonary injury† | 505 (49.0%) | 321 (71.5%) | <0.001 |
| Comorbidity, n (%) | |||
| Diabetes | 282 (27.3%) | 78 (17.4%) | <0.001 |
| Liver cirrhosis/failure | 39 (3. 8%) | 31 (6.9%) | 0.009 |
| Corticosteroid treatment before ICU admission, n (%)‡ | 93 (9.0%) | 47 (10.5%) | 0.381 |
| History of alcohol abuse |
101 (9.8%) |
64 (14.2%) |
0.012 |
Definition of abbreviations: ARDS, acute respiratory distress syndrome; APACHE, Acute Physiology and Chronic Health Evaluation.
APACHE III physiology score was calculated with all components on the day of ICU admission.
Pneumonia, aspiration, or pulmonary contusions are categorized as pulmonary injury. Sepsis from an extrapulmonary source, trauma without pulmonary contusions, and multiple transfusions were categorized as extrapulmonary injury. Patients with both pulmonary and extrapulmonary injury were considered to have pulmonary injury.
Patient received ≥ 300 mg of prednisone or its equivalent within 21 d or ≥ 15 mg prednisone a day or its equivalent before ICU admission.
Genetic Variations at PI3 Locus
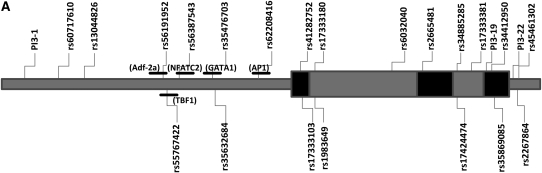
We identified a total of 24 polymorphisms at PI3 locus, including 21 SNPs and 3 deletion/insertion polymorphisms. Among them, three polymorphisms were novel variants that were neither published in dbSNP (http://www.ncbi.nlm.nih.gov/SNP), nor reported elsewhere. Positions and allelic frequencies of the identified SNPs are shown in Table 2. We identified nine polymorphisms in the promoter region of PI3. Most of them have been predicted to have different binding sites for one or more transcription factors (27). Three nonsynonymous polymorphisms, resulting in amino acid substitutions and previously described in the dbSNP database, were found in the coding regions of the gene. In addition, there were two novel SNPs (PI3–19 and PI3–22) identified in the 3′-untranslated region. The genomic structure of PI3 and the relative physical position of the polymorphisms are presented in Figure 1A.
TABLE 2.
POLYMORPHISMS DETECTED IN PI3 GENE
| No* | SNP ID | Chr20 nt position† | Region | Alleles | MAF (n)‡ |
|---|---|---|---|---|---|
| PI3-1 | Novel | 43234809 | Promoter | T>A | 0.136 (44) |
| PI3-2 | rs60717610 | 43235071 | Promoter | –/TAATA | 0.214 (56) |
| PI3-3 | rs13044826 | 43235279 | Promoter | G>A | 0.214 (56) |
| PI3-4 | rs56191952 | 43235901 | Promoter | A>G | 0.214 (56) |
| PI3-5 | rs55767422 | 43235911 | Promoter | A>G | 0.214 (56) |
| PI3-6 | rs56387543 | 43236018 | Promoter | –/T | 0.214 (56) |
| PI3-7 | rs35476703 | 43236289 | Promoter | C>G | 0.211 (52) |
| PI3-8 | rs35632684 | 43236303 | Promoter | C>T | 0.211 (52) |
| PI3-9 | rs62208416 | 43236640 | Promoter | G>A | 0.214 (56) |
| PI3-10 | rs41282752 | 43237020 | Exon 1 | G>A, A15T | 0.018 (56) |
| PI3-11 | rs17333103 | 43237027 | Exon 1 | C>T, T17M | 0.232 (56) |
| PI3-12 | rs17333180 | 43237122 | intron 1 | C>A | 0.214 (56) |
| PI3-13 | rs1983649 | 43237139 | intron 1 | A>T | 0.340 (56) |
| PI3-14 | rs6032040 | 43237728 | intron 1 | T>A | 0.108 (56) |
| PI3-15 | rs2664581 | 43237936 | Exon 2 | A>C, T34P | 0.203 (54) |
| PI3-16 | rs34885285 | 43238203 | intron 2 | C>A | 0.203 (54) |
| PI3-17 | rs17424474 | 43238211 | intron 2 | C>A | 0.203 (54) |
| PI3-18 | rs17333381 | 43238333 | intron 2 | C>T | 0.203 (54) |
| PI3-19 | Novel | 43238432 | Exon 3 (3′UTR) | C>A | 0.018 (56) |
| PI3-20 | rs34412950 | 43238546 | Exon 3 (3′UTR) | C>A | 0.214 (56) |
| PI3-21 | rs35869085 | 43238548 | Exon 3 (3′UTR) | G>C | 0.214 (56) |
| PI3-22 | Novel | 43238671 | 3′UTR | –/TCT | 0.214 (56) |
| PI3-23 | rs45461302 | 43238688 | 3′UTR | G>A | 0.196 (56) |
| PI3-24 |
rs2267864 |
43238693 |
3′UTR |
C>G |
0.203 (54) |
Definition of abbreviations: MAF, minor allele frequency; SNP, single-nucleotide polymorphism; nt, nucleotide; UTR, untranslated region.
Polymorphisms are presented in chromosomal order and their position within the gene is indicated.
Chromosome position based on National Center for Biotechnology Information.
(n) is the number of chromosomes used in the estimation of MAF.
Figure 1.
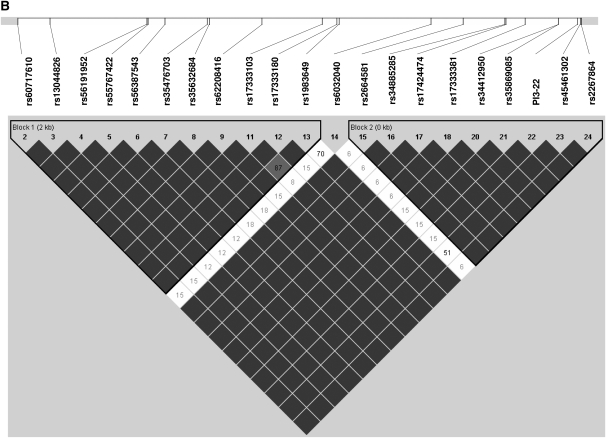
(A) Location of the single-nucleotide polymorphisms (SNPs) identified in peptidase inhibitor (PI3) gene. The first nucleotide of the translation start site is denoted as +1. Dark shaded and light shaded boxes represent exons and introns, respectively. Horizontal bars indicate the transcription factor predicted to bind to one of the alleles of rs56191952, rs55767422, rs56387543, rs35476703, and rs62208416, respectively (20). (B) Linkage disequilibrium plot for SNPs in the PI3 gene. Numbers within each diamond are D′ between pairs of SNPs expressed in percentages. The black-to-white gradient reflects higher to lower linkage disequilibrium values (black, high; white, low). Unreported values reflect D′ = 1 (100%).
We used resequencing data to determine the LD structure of the PI3 gene (Figure 1B). Two LD blocks were found that were constructed with SNPs rs60717610-rs1983649 and rs2664581-rs2267864. Each block showed limited haplotype diversity, with high coefficient between the two LD blocks (D′= 1). We identified three tSNPs, rs19836491, rs6032040, and rs2664581 (PI3–13, PI3–14, and PI3–15, respectively), which capture all variants with MAF ≥ 10%.
Genotype distributions of the selected tSNPs in the at-risk controls were in Hardy-Weinberg equilibrium. MAFs for tSNPs between cases and control subjects are shown in Table 3. We found a significant difference in MAF of rs2664581 between the two groups (P = 0.008, FDR adjusted P = 0.024). We also found a significant difference in the genotype distribution of rs2664581 between patients with ARDS and control subjects, but this difference was not statistically significant after adjustment for multiple comparisons (P = 0.030, FDR adjusted P = 0.090) (Table E3).
TABLE 3.
SNPS GENOTYPED IN THE CURRENT STUDY
| Minor Allele Frequency |
||||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Allele | Location | Chr20 nt position* | All | ARDS | Control | P Value† | P Value‡ |
| rs1983649 | A>T | intron 1 | 43237139 | 0.417 | 0.429 | 0.412 | 0.389 | 0.800 |
| rs6032040 | T>A | intron 1 | 43237728 | 0.170 | 0.161 | 0.173 | 0.456 | 0.183 |
| rs2664581 |
A>C |
exon 2 |
43237936 |
0.183 |
0.214 |
0.173 |
0.008 |
0.740 |
Definition of abbreviations: ARDS, acute respiratory distress syndrome; nt, nucleotide; SNP, single-nucleotide polymorphism.
Chromosome position based on National Center for Biotechnology Information.
The probability of the chi-square test for comparison of allele frequencies.
The probability of the chi-square test for deviation from Hardy-Weinberg equilibrium in the control group.
Association between PI3 Polymorphisms and the Risk of Developing ARDS
Table 4 summarizes the results of association tests assuming additive and dominant models. Under the additive model, the minor allele of rs2664581 (C allele) was significantly associated with increased risk of developing ARDS in both crude (OR, 1.30; 95% CI, 1.07–1.58; P = 0.008; FDR adjusted P = 0.025) and adjusted analyses (OR, 1.35; 95% CI, 1.09–1.67; P = 0.006; FDR adjusted P = 0.018). In stratified analysis by source of lung injury, the minor alleles of rs2664581 (C allele) and rs1983649 (T allele) were significantly associated with increased ARDS risk in patients with extrapulmonary injury, whereas no association with ARDS risk was found among patients with pulmonary injury. When we tested the association using a dominant genetic model, we obtained similar results that C allele of rs2664581 was strongly associated with increased risk of developing ARDS (Table 4).
TABLE 4.
ASSOCIATION BETWEEN PI3 GENOTYPES AND ACUTE RESPIRATORY DISTRESS SYNDROME RISK
| All Subjects (n = 1,480) |
Pulmonary Injury (n = 831) |
Extrapulmonary Injury (n = 649) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Crude OR (95% CI) | Adjusted OR‡ (95% CI) | P Value | Crude OR (95% CI) | Adjusted OR‡ (95% CI) | P Value | Crude OR (95% CI) | Adjusted OR‡ (95% CI) | P Value |
| rs1983649 | |||||||||
| Additive* | 1.07 (0.91–1.25) | 1.09 (0.92–1.29) | 0.324 | 0.97 (0.79–1.18) | 0.95 (0.77–1.18) | 0.635 | 1.29 (0.98–1.70) | 1.37 (1.02–1.84) | 0.034‖ |
| Dominant† | 1.08 (0.85–1.36) | 1.10 (0.85–1.42) | 0.449 | 0.97 (0.73–1.31) | 0.98 (0.71–1.34) | 0.880 | 1.26 (0.83–1.92) | 1.35 (0.86–2.10) | 0.187 |
| rs6032040 | |||||||||
| Additive* | 0.92 (0.75–1.13) | 0.96 (0.77–1.21) | 0.759 | 0.83 (0.64–1.07) | 0.86 (0.65–1.14) | 0.303 | 1.08 (0.75–1.57) | 1.09 (0.73–1.61) | 0.670 |
| Dominant† | 1.48 (0.75–2.93) | 1.45 (0.70–2.98) | 0.317 | 2.27 (0.96–5.34) | 1.95 (0.80–4.78) | 0.144 | 0.75 (0.24–2.35) | 0.85 (0.24–2.96) | 0.797 |
| rs2664581 | |||||||||
| Additive* | 1.30 (1.07–1.58) | 1.35 (1.09–1.67) | 0.006§ | 1.24 (0.97–1.60) | 1.19 (0.91–1.58) | 0.193 | 1.48 (1.06–2.06) | 1.65 (1.16–2.34) | 0.004¶ |
| Dominant† |
1.35 (1.07–1.70) |
1.43 (1.11–1.83) |
0.006§ |
1.34 (1.00–1.80) |
1.32 (0.96–1.81) |
0.082 |
1.42 (0.94–2.13) |
1.64 (1.07–2.52) |
0.023‖ |
Definition of abbreviations: CI, confidence interval; OR, odds ratio; SNP, single nucleotide polymorphism.
* In the additive model, OR were expressed per difference in the number of minor alleles (T allele of rs1983649; A allele of rs603240; and C allele of 2664581).
In the dominant model, OR were expressed as heterozygotes and rare homozygotes compared with common homozygotes (AT/TT versus AA for rs1983649; TA/AA versus TT for rs6032040; and AC/CC versus AA for 2664581).
Adjusted for age, sex, APACHE III score, risk factors for ARDS, comorbidities (diabetes, liver cirrhosis/failure), and alcohol abuse.
FDR P = 0.018.
FDR P = 0.051.
FDR P = 0.012.
Eight inferred haplotypes were observed in the studied population. Four of them occurred at frequencies over 5% and jointly accounted for a cumulative frequency of 99.8% in our population (Table 5). Significant differences were observed in the distribution of Hap2 (TTC) between ARDS and control subjects (P = 0.009, FDR adjusted P = 0.036).
TABLE 5.
ASSOCIATION BETWEEN PI3 HAPLOTYPES AND ACUTE RESPIRATORY DISTRESS SYNDROME RISK
| All subjects (n = 1480) |
Pulmonary injury (n = 831) |
Extrapulmonary injury (n = 649) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Haplotype Identification | Haplotype Composition* | Haplotype Frequency (ARDS/Controls) | Adjusted OR (95% CI)† | P Value | Haplotype Frequency (ARDS/Controls) | Adjusted OR (95% CI)† | P Value | Haplotype Frequency (ARDS/Controls) | Adjusted OR (95% CI)† | P Value |
| Global test‡ | 0.060 | 0.373 | 0.096 | |||||||
| Hap1 | ATA (0-0-0) | 56.8/58.7 | 1.0 | — | 58.3/57.5 | 1.0 | — | 52.9/60.0 | 1.0 | — |
| Hap2 | TTC (1-0-1) | 21.2/17.2§ | 1.31 (1.05–1.64) | 0.015 | 20.2/17.0 | 1.14 (0.86–1.51) | 0.370 | 23.9/17.3 | 1.67 (1.16–2.40) | 0.005 |
| Hap3 | TAA (1-1-0) | 17.2/15.9 | 0.99 (0.78–1.26) | 0.096 | 15.8/18.6 | 0.87 (0.65–1.16) | 0.353 | 16.2/15.9 | 1.17 (0.77–1.79) | 0.447 |
| Hap4 | TTA (1-0-0) | 5.7/6.7 | 0.78 (0.54–1.13) | 0.185 | 5.5/6.7 | 0.72 (0.46–1.14) | 0.167 | 6.1/6.7 | 0.97(0.51–1.79) | 0.895 |
| Others‖ |
— |
— |
— |
— |
— |
— |
— |
— |
— |
— |
Definition of abbreviations: ARDS, acute respiratory distress syndrome; CI, confidence interval; OR, odds ratio.
Individual tSNPs alleles are presented in chromosomal order rs1983649, rs6032040, and rs2664581.
Adjusted for age, sex, APACHE III score, risk factors for ARDS, comorbidities, alcohol abuse, recent use of steroid.
Global test for haplotypes by likehood-ratio test (LRT), with 4 degrees of freedom.
Chi-square test for comparison of haplotypes frequencies between patients with ARDS and control subjects was P = 0.009, FDR P = 0.036.
Haplotypes with frequency < 0.01%, were not included in the analysis.
Haplotype-specific risk for ARDS was assessed using the most common haplotype (Hap1: ATA) as the reference in the regression model. The global haplotype association test demonstrated that PI3 haplotypes were marginally associated with ARDS risk (LRT, P = 0.06). Consistent with the results of single polymorphism analysis, the haplotype (Hap2: TTC) containing variant alleles of rs2664581 and rs1983649 was significantly associated with increased ARDS risk (OR, 1.31; 95% CI, 1.05–1.64; P = 0.015). In the stratified analysis, the same haplotype was associated with risk of ARDS among patients with extrapulmonary injury (OR, 1.67; 95% CI, 1.16–2.40; P = 0.005), but the global test was not significant in this subgroup (LRT, P = 0.096) (Table 5).
Association between PI3 Polymorphisms and ARDS Survival
Baseline characteristics between survivors and nonsurvivors among patients with ARDS are shown in Table E4. Among patients with ARDS, the survivors were younger and had lower APACHE III scores than the nonsurvivors. There were no significant differences in mortality rates at either 28 days or 60 days between survivors and nonsurvivors for any selected tSNP. None of the PI3 genotypes or haplotypes was significantly associated with 60-day survival in all subjects or in stratified subgroups (Table E5).
PI3 Polymorphisms and Plasma PI3 Levels
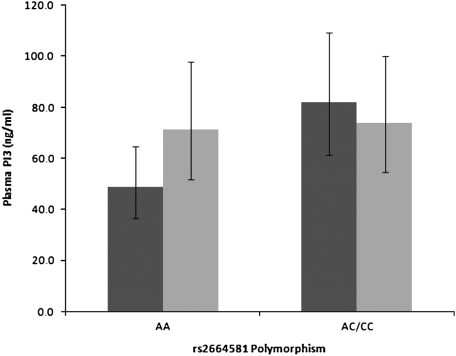
We investigated the relationship of rs2664581 polymorphism and plasma PI3 level. As shown in Figure 2, patients with ARDS with C allele (variant allele corresponding to P residue, n = 19; mean = 81.9 ng/ml; 95% CI, 61.3–109.4 ng/ml) had a significant higher level of plasma PI3 at ARDS onset as compared with those patients with ARDS with homozygous A allele (wild-type allele; n = 38; mean = 48.6 ng/ml; 95% CI, 36.6–64.6 ng/ml) in both two sample t test (P = 0.019) and general linear model analysis (P = 0.043) adjusted by age, sex, ethnicity, type of lung injury, pre-admission steroid use, septic shock, and APACHE III score. In contrast, there was no difference between different alleles of rs2664581 in control subjects (C allele, n = 21, mean = 74.0 ng/ml; 95% CI, 54.7–100.1 ng/ml; and A allele, n = 35, mean = 71.1 ng/ml; 95% CI, 51.8–97.7 ng/ml) (two sample t-test, P = 0.868; GLM, P = 0.121). Moreover, in the subgroup analysis of patients homozygous for A allele, the patients with ARDS had significant lower plasma PI3 than the control subjects (two sample t test, P = 0.077; GLM, P = 0.024). On the contrary, among patients carrying variant allele, we did not observe any difference of plasma profiles between the patients with ARDS and the control subjects (two sample t-test, P = 0.617; GLM, P = 0.107). ANOVA analyses demonstrated there were significant differences among four subgroups (genotypes by ARDS status) after adjustment of age, sex, ethnicity, pulmonary injury, pre-admission steroid use, septic shock, and APACHE III score on ICU admission (P = 0.003). Furthermore, in pairwise comparison with correction for multiple comparison (Tukey test), we still found significant higher plasma PI3 in patients with ARDS with C allele as compared with patients with ARDS with homozygous A allele.
Figure 2.
Effects of PI3 rs2664581 polymorphism on plasma PI3 level. The acute respiratoyr distress syndrome (ARDS) samples were collected within 48 hours of ARDS diagnosis, and the control samples were collected within 48 hours of ICU admission. Geometric mean of plasma PI3 is shown in columns with error bar representing 95% confidence interval. Since the natural log transformed data were used in the analyses, antilogs of the means and 95% confidence intervals are presented in the figure. The subgroup of the ARDS with AA genotype (n = 35) had significant lower plasma PI3 than the ARDS with AC/CC genotypes (n = 19, P = 0.043) and the control with AA genotype (n = 38, P = 0.121) in GLM analyses. P values after adjustment for age, sex, type of lung injury, pre-admission steroid use, septic shock, and APACHE III score. Dark shaded bars, ARDS; light shaded bars, control.
Among patients with pulmonary injury, control subjects had significantly higher PI3 plasma levels (n = 29; mean = 85.5 ng/ml; 95% CI, 59.4–123.1 ng/ml) compared with patients with ARDS (n = 44; mean = 55.8 ng/ml; 95% CI, 43.3–71.8 ng/ml) (two sample t-test, P = 0.047; GLM, P = 0.008). We did not observe significant difference in plasma PI3 levels between ARDS cases and control subjects in the subgroup of patients with extrapulmonary injury (two sample t test, P = 0.566), given a relatively small number of cases (n = 10) in contrast to control subjects (n = 31). Moreover, there were no significant differences in PI3 levels between pulmonary versus extrapulmonary injury among ARDS cases or control subjects (two sample t test, P = 0.371 and 0.146, respectively). We did not conduct further stratified analysis to investigate the association between genotypes and plasma PI3 level due to the limitation of small sample size in subgroups with variant allele of rs2664581.
DISCUSSION
In the present study, we identified 24 polymorphisms, including 3 novel variants, in PI3 gene within the studied non-Hispanic white population. Three tSNPs (rs1983649, rs6032040, and rs2664581), which capture the entire common genetic variations in PI3 gene, were genotyped and analyzed for the association with ARDS. We found that the variant allele of rs2664581 was significantly associated with increased ARDS risk. The haplotype (Hap2: TTC), which contains variant C allele of rs2664581, was also associated with increased risk for ARDS. However, we did not find any significant associations between PI3 polymorphisms and all-cause 60-day mortalities. Thus, our results support the hypothesis that genetic variations in the PI3 gene are associated with major susceptibility to ARDS.
In addition, we found that the genetic effects of PI3 polymorphisms on ARDS development were modified by the origin of lung injury. In stratified analysis, variants of rs1983649 and rs2664581 were individually associated with increased ARDS risk in patients with extrapulmonary injury. Consistent with the SNP analysis, Hap2 containing the variant alleles of rs1983649 and rs2664581 was also associated with ARDS development in patients with extrapulmonary injury. However, this association was not observed in patients with Hap4 carrying the wild-type allele of rs2664581, but only variant allele of rs1983649, suggesting that the association between rs1983649 and ARDS is driven by LD with rs2664581.
The modification of the genetic association with ARDS by the origin of lung injury has been observed previously. Recent studies in our cohort indicated that the −308G/A and LTA polymorphisms of the tumor necrosis factor genes (TNFB) were associated with ARDS in patients with pulmonary, but not extrapulmonary injury (16). In the same cohort women with the variant SFTPB polymorphism of surfactant protein-B gene (SP-B) had an increased odd of having a pulmonary injury as a risk factor for ARDS (15). These results were consistent with a recent report in which the −1580C/T missense mutation in the SP-B gene was associated to increased risk of ARDS in patients with pulmonary injury (28). It is known that ARDS derived from a pulmonary insult has different pathophysiologic, biochemical, radiologic, and mechanical patterns from ARDS caused by an extrapulmonary injury (29). In ARDS derived from a pulmonary insult (pulmonary ARDS), the prevalent damage in early stage is likely intra-alveolar, whereas in ARDS caused by extrapulmonary insult (extrapulmonary ARDS) the damage is focused on the vascular endothelium with a greater amount of inflammatory agents in the bloodstream. Recently, it has been also demonstrated that acute lung injury derived from different insults leads to different gene expression profiles. Genes stimulated by lipopolysaccharide in animal model from extrapulmonary acute lung injury were involved in metabolism, defense response, immune cell proliferation, differentiation and migration, and cell death, whereas genes involved in organogenesis, morphogenesis, cell cycle, proliferation, and differentiation were significantly expressed in response to high-volume ventilation (30). Our results indicate that genetic variants of rs2664581 may contribute to those processes involved in the pathogenesis of extrapulmonary ARDS rather than those involved in the pathogenesis of pulmonary ARDS. HNE has been shown to compromise the integrity of the endothelial vascular barrier and promote microvascular injury during ARDS development (4, 31). Variants of rs2664581 (or other SNPs in LD) could affect the capability of PI3 to inhibit the elevated HNE activity and therefore increase the risk for developing ARDS, principally among patients with extrapulmonary injury. However, given the reduced sample size in the subgroups, our findings will need to be confirmed and explored in a larger population with clinically homogeneous risks for ARDS.
We also found that, at the onset of ARDS, plasma PI3 levels were affected by different genotypes of rs2664581 (T34P) polymorphism. Patients with ARDS homozygous for A allele had a significant lower PI3 plasma levels than those with variant C allele. Moreover, among patients homozygous for A allele, ARDS cases had significant lower plasma PI3 levels as compared with the at-risk control subjects. Our previous studies showed that PI3 might play a protective role against ARDS development. However, the C allele in rs2665481 associated with higher odds of developing ARDS is also associated with higher plasma PI3 in patients with ARDS. A possible explanation for this apparent contradiction is that the amino acid substitution associated with the C allele results in a defective protein, and a higher level of defective PI3 protein could not provide enough protection against deleterious effects of excessive HNE.
The SNP rs2664581 is a nonsynonymous polymorphism located in exon 2 that results in an amino acid substitution (T→P) within the consensus sequence GQDPVK of the first transglutaminase substrate motif of PI3 (11). The capability of PI3 to bind covalently to the ECM through transglutaminase substrate motifs and exert its biological function make it effective as a tissue-bound inhibitor against HNE (12). Although the substitution T→P is located at a nonconservative position of transglutaminase substrate motifs across different protein families, and the second and fifth transglutaminase substrate motifs of PI3 had the same proline at this position, a threonine at this position (amino acid 34) is well conserved in elafins across all species (Figure E2). Moreover, the first transglutaminase substrate motif is close to a signal peptide cleavage site so that this motif may be more easily accessed to transglutaminase (9, 11). Thus, this motif may be more important than the rest of the transglutaminase motifs in binding to ECM proteins, and is more sensitive to proline substitution. Proline is known to break a helix of three-dimensional protein structure with significant conformational changes. It is possible that the structure disruption caused by the T34P substitution affects the ability of the PI3 protein to stay bound to ECM protein or else increases the ability of pre-elafin to be cleaved into elafin, which results in a reduction of effective local protection by increasing free PI3 in local tissue and/or circulation.
Recently, we conducted a comprehensive investigation of plasma profiles of PI3, SLPI, and HNE in patients with ARDS and at-risk, critically ill control subjects (32). We found that the patients with ARDS had significantly higher plasma HNE but lower PI3, resulting in increased HNE/PI3 ratio at the onset of ARDS, compared with at-risk control subjects at the ICU hospitalization, who had significant increase of both HNE and PI3, keeping a normal HNE/PI3 ratio as compared with healthy individuals. Before the diagnosis of ARDS, we observed that patients with ARDS had plasma profiles similar to those of control subjects with both elevated plasma HNE and PI3. Along with the progress of acute lung injury to the onset of ARDS, there was a decrease of plasma PI3 levels, whereas HNE was maintained at a higher level, characterized by increased HNE/PI3 ratio. In contrast, there was a significant drop of the HNE/PI3 ratio in the at-risk control subjects 3 days after ICU admission. Therefore, the loss of the balance between plasma HNE and its specific inhibitor PI3 toward more neutrophil elastase is related to the risk of ARDS development. More specifically, the decrease PI3 along with the clinical progress of ARDS might be the cause for disrupting the HNE-PI3 balance. These findings agree with our hypothesis of defective PI3 protein encoded by rs2664581 (T34P) polymorphism. At the onset of ARDS, HNE/PI3 ratio might be normal but the variant PI3 with proline substitute could not provide enough protection.
The results described above seemed to point a nonsynonymous polymorphism rs2664581 (T34P) as a putative causal SNP in ARDS development. The haplotype analysis showed that rs2665481 is independently associated with increased ARDS risk. However, the LD structure around PI3 gene complicates the identification of rs2665481 as the causal SNP able to modify the risk of ARDS (Figure 1B). Similar to a previous study in an African-American population (27), we found the PI3 gene in our studied population was also highly polymorphic. All polymorphisms were located within LD blocks with high degree of linkage disequilibrium (Figure 1B). There were nine polymorphisms located in the promoter region, with potential differential binding for at least one transcription factor, including Adf-2a, TBF1, NFATC2, AP1, and GATA1 (27, 33–35). Most interestingly, all of them were in complete linkage disequilibrium with rs2664581 (T34P) polymorphism. It is possible that PI3 plasma levels could be influenced by SNPs in the promoter region, which could modify gene transcription by altering the binding of transcription factors to promoter or by altering the local chromatin architecture. Chowdhury and coworkers identified two SNPs in the PI3 promoter (−1063G→A, not identified in our population, and rs35476703) which showed differential binding of transcription factors in nuclear extracts derived from both amnion and HeLa cells (27). The SNP rs35476703 site at −689C→G binds to GATA1, suggesting the involvement of this transcription factor in regulation of PI3 in amnion cells. We did not genotype this SNP because it is not a tSNP based on disequilibrium criteria, and genotyping this SNP would not provide additional information because they are essentially in complete LD with SNP rs2664581 (T34P). SNPs in 3′-untranslated region of the PI3 gene, in high LD with rs2664581, could also alter message stability. Furthermore, it is also possible that the functional SNPs could be located outside the PI3 locus in LD with tSNP rs2664581 (T34P).
Taking all together, it is likely that PI3 polymorphisms are acting synergically with multiple effects at transcriptional, post-transcriptional, and post-translational levels, which might play important roles in the ARDS development. Nevertheless, our conclusions are based exclusively in the analysis of the correlation genotype–intermediate phenotype (rs2664581 genotypes/plasma PI3 levels). Biological studies are still required to investigate the functional significance of polymorphisms in PI3 gene and to let us make a causal inference.
To our knowledge, this is the first study to examine the role of genetic variations in PI3 gene in the predisposition to ARDS. Our study hypothesis was based on the biological plausibility that PI3 functioned as a specific inhibitor of HNE (9), and was supported by our previous findings that pointed to PI3 gene as a candidate for ARDS susceptibility (14). We applied several approaches to reduce potential selection biases and confounding factors in the hospital-based study design. First, the cases were strictly defined according to AECC definition for ARDS (18). Second, the selection of controls from at-risk critically ill patients reduced the potential confounding from associations between candidate polymorphisms and predisposing conditions for ARDS. Third, all subjects in this study belonged to a single ethnic group, thus the potential confounding effect due to population stratification is minimized. In addition, this study included 449 ARDS cases and 1,031 control subjects, representing the largest population used in an ARDS association study, and provided high statistical power to detect genetic effects on ARDS. The study also had a complete coverage of all common polymorphisms of PI3 gene to ensure the comprehensive evaluation of the association of PI3 polymorphisms with ARDS risk. The tSNPs selection was based on our resequencing data that captured all common variants surrounding the PI3 gene with an r2 ≥ 0.8, and was consistent with those predicted by the HapMap data of CEU sample (release 22, on NCBI B36, dbSNP b126. http://www.hapmap.org/cgi-perl/gbrowse/hapmap22_B36) (Figure E3).
We acknowledge some limitations in our study. Since we included only non-Hispanic white patients, the results cannot be extrapolated directly to other ethnic groups. In addition, the use of samples from other ethnic groups with different LD structures could help to better characterize the identification of SNPs that have a functional role in ARDS development (36), as we found different LD strucutures of PI3 gene among different populations using HapMap data of CHB, JPT, YRI, and CEU samples. The LD pattern identified in our population was well correlated with that predicted by the HapMap in white populations. On the other hand, although our study has been restricted to non-Hispanic white subjects, the ancestry of the participants has not been genetically determined. Controversy exists regarding whether self-reported ancestry accounts for major axes of ancestry variation in the European-American population (37, 38). As such, we cannot exclude the possibility of population stratification in our study. However, population stratification is the result of both differences in allele frequency and disease risk. These interethnic differences in disease risk are determined not only genetically but also by differences in cultural and behavioral factors. Self-reported ancestry is more related to these factors than genome-based ethnicity (37). Although the genomic determination of ethnicity cannot be inferred from self-reported ancestry, the inclusion of self-reported ancestry in our study allows us to control the confounding by interethnic differences in ARDS risk.
Although we report on the largest population in an ARDS association study, the number of ARDS cases with available plasma samples was small. In the stratified analysis by type of lung injury, we found that control subjects had a higher plasma PI3 level than ARDS in the pulmonary injury group. According to the genotype analysis, the genetic association with ARDS is driven by extrapulmonary injury. However, no differences in plasma PI3 level between patients with ARDS and control subjects were found among the extrapulmonary injury group. The small size of the group of patients with ARDS with extrapulmonary injury (n = 10) could be a possible explanation for the lack of association in this group. Finally, our findings need be replicated in other populations.
In summary, our results demonstrate a significant association between PI3 polymorphisms and risk for ARDS. The SNPs in PI3 gene may be important in controlling PI3 plasma levels and the capability to protect from HNE-induced lung injury. Our study provides rationale for more detailed functional characterization of the SNPs in PI3 gene, which will enable us to understand the role of PI3 in ARDS pathogenesis.
Acknowledgments
The authors thank Weiling Zhang, Kelly McCoy, Thomas McCabe, Julia Shin, for patient recruitment; Andrea Shafer and Lia Shimada for research support; Maureen Convery for laboratory expertise; and Janna Frelich and Marcia Chertok for data management.
This work was supported by National Institutes of Health grants HL 060710 and ES00002. P.T. was supported by grant from Fundación Ramón Areces (Spain).
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2008-0410OC on February 27, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Bellingan GJ. The pulmonary physician in critical care * 6: the pathogenesis of ALI/ARDS. Thorax 2002;57:540–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ware LB, Matthay MA. The acute respiratory distress syndrome. N Engl J Med 2000;342:1334–1349. [DOI] [PubMed] [Google Scholar]
- 3.Weiland JE, Davis WB, Holter JF, Mohammed JR, Dorinsky PM, Gadek JE. Lung neutrophils in the adult respiratory distress syndrome: clinical and pathophysiologic significance. Am Rev Respir Dis 1986;133:218–225. [DOI] [PubMed] [Google Scholar]
- 4.Kawabata K, Hagio T, Matsuoka S. The role of neutrophil elastase in acute lung injury. Eur J Pharmacol 2002;451:1–10. [DOI] [PubMed] [Google Scholar]
- 5.Pfundt R, Wingens M, Bergers M, Zweers M, Frenken M, Schalkwijk J. TNF-alpha and serum induce SKALP/elafin gene expression in human keratinocytes by a p38 MAP kinase-dependent pathway. Arch Dermatol Res 2000;292:180–187. [DOI] [PubMed] [Google Scholar]
- 6.Reid PT, Marsden ME, Cunningham GA, Haslett C, Sallenave JM. Human neutrophil elastase regulates the expression and secretion of elafin (elastase-specific inhibitor) in type II alveolar epithelial cells. FEBS Lett 1999;457:33–37. [DOI] [PubMed] [Google Scholar]
- 7.Sallenave JM, Shulmann J, Crossley J, Jordana M, Gauldie J. Regulation of secretory leukocyte proteinase inhibitor (SLPI) and elastase-specific inhibitor (ESI/elafin) in human airway epithelial cells by cytokines and neutrophilic enzymes. Am J Respir Cell Mol Biol 1994;11:733–741. [DOI] [PubMed] [Google Scholar]
- 8.Pfundt R, van Ruissen F, van Vlijmen-Willems IM, Alkemade HA, Zeeuwen PL, Jap PH, Dijkman H, Fransen J, Croes H, van Erp PE, et al. Constitutive and inducible expression of SKALP/elafin provides anti-elastase defense in human epithelia. J Clin Invest 1996;98:1389–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreau T, Baranger K, Dade S, Dallet-Choisy S, Guyot N, Zani ML. Multifaceted roles of human elafin and secretory leukocyte proteinase inhibitor (SLPI), two serine protease inhibitors of the chelonianin family. Biochimie 2008;90:284–295. [DOI] [PubMed] [Google Scholar]
- 10.Nara K, Ito S, Ito T, Suzuki Y, Ghoneim MA, Tachibana S, Hirose S. Elastase inhibitor elafin is a new type of proteinase inhibitor which has a transglutaminase-mediated anchoring sequence termed “cementoin”. J Biochem 1994;115:441–448. (Tokyo). [DOI] [PubMed] [Google Scholar]
- 11.Schalkwijk J, Wiedow O, Hirose S. The trappin gene family: proteins defined by an N-terminal transglutaminase substrate domain and a C-terminal four-disulphide core. Biochem J 1999;34:569–577. [PMC free article] [PubMed] [Google Scholar]
- 12.Guyot N, Zani ML, Maurel MC, Dallet-Choisy S, Moreau T. Elafin and its precursor trappin-2 still inhibit neutrophil serine proteinases when they are covalently bound to extracellular matrix proteins by tissue transglutaminase. Biochemistry 2005;44:15610–15618. [DOI] [PubMed] [Google Scholar]
- 13.Williams SE, Brown TI, Roghanian A, Sallenave JM. SLPI and elafin: one glove, many fingers. Clin Sci (Lond) 2006;110:21–35. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Beach D, Su L, Zhai R, Christiani DC. A genome-wide expression analysis in blood identifies pre-elafin as a biomarker in ARDS. Am J Respir Cell Mol Biol 2008;38:724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gong MN, Wei Z, Xu LL, Miller DP, Thompson BT, Christiani DC. Polymorphism in the surfactant protein-B gene, gender, and the risk of direct pulmonary injury and ARDS. Chest 2004;125:203–211. [DOI] [PubMed] [Google Scholar]
- 16.Gong MN, Zhou W, Williams PL, Thompson BT, Pothier L, Boyce P, Christiani DC. -308GA and TNFB polymorphisms in acute respiratory distress syndrome. Eur Respir J 2005;26:382–389. [DOI] [PubMed] [Google Scholar]
- 17.Gong MN, Thompson BT, Williams P, Pothier L, Boyce PD, Christiani DC. Clinical predictors of and mortality in acute respiratory distress syndrome: potential role of red cell transfusion. Crit Care Med 2005;33:1191–1198. [DOI] [PubMed] [Google Scholar]
- 18.Bernard GR, Artigas A, Brigham KL, Carlet J, Falke K, Hudson L, Lamy M, Legall JR, Morris A, Spragg R. The American-European Consensus Conference on ARDS: definitions, mechanisms, relevant outcomes, and clinical trial coordination. Am J Respir Crit Care Med 1994;149:818–824. [DOI] [PubMed] [Google Scholar]
- 19.Sallenave JM, Silva A. Characterization and gene sequence of the precursor of elafin, an elastase-specific inhibitor in bronchial secretions. Am J Respir Cell Mol Biol 1993;8:439–445. [DOI] [PubMed] [Google Scholar]
- 20.Kruglyak L, Nickerson DA. Variation is the spice of life. Nat Genet 2001;27:234–236. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005;21:263–265. [DOI] [PubMed] [Google Scholar]
- 22.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. The structure of haplotype blocks in the human genome. Science 2002;296:2225–2229. [DOI] [PubMed] [Google Scholar]
- 23.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet 2005;37:1217–1223. [DOI] [PubMed] [Google Scholar]
- 24.Kraft P, Cox DG, Paynter RA, Hunter D, De Vivo I. Accounting for haplotype uncertainty in matched association studies: a comparison of simple and flexible techniques. Genet Epidemiol 2005;28:261–272. [DOI] [PubMed] [Google Scholar]
- 25.Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered 2002;53:79–91. [DOI] [PubMed] [Google Scholar]
- 26.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res 2001;125:279–284. [DOI] [PubMed] [Google Scholar]
- 27.Chowdhury MA, Kuivaniemi H, Romero R, Edwin S, Chaiworapongsa T, Tromp G. Identification of novel functional sequence variants in the gene for peptidase inhibitor 3. BMC Med Genet 2006;7:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Z, Pearson C, Chinchilli V, Pietschmann SM, Luo J, Pison Y, Floros J. Polymorphisms of human SP-A, SP-B, and SP-D genes: association of SP-B Thr131Ile with ARDS. Clin Genet 2000;58:181–191. [DOI] [PubMed] [Google Scholar]
- 29.Pelosi P, D'Onofrio D, Chiumello D, Paolo S, Chiara G, Capelozzi VL, Barbas CS, Chiaranda M, Gattinoni L. Pulmonary and extrapulmonary acute respiratory distress syndrome are different. Eur Respir J Suppl 2003;42:48s–56s. [DOI] [PubMed] [Google Scholar]
- 30.dos Santos CC, Okutani D, Hu P, Han B, Crimi E, He X, Keshavjee S, Greenwood C, Slutsky AS, Zhang H, et al. Differential gene profiling in acute lung injury identifies injury-specific gene expression. Crit Care Med 2008;36:855–865. [DOI] [PubMed] [Google Scholar]
- 31.Moraes TJ, Chow CW, Downey GP. Proteases and lung injury. Crit Care Med 2003;31:S189–S194. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Chen F, Zhai R, Zhang L, Su L, Lin X, Thompson T, Christiani DC. Plasma neutrophil elastase and elafin imbalance is associated with acute respiratory distress syndrome (ARDS) development. PLoS One 2009;4:e4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bingle L, Tetley TD, Bingle CD. Cytokine-mediated induction of the human elafin gene in pulmonary epithelial cells is regulated by nuclear factor-kappaB. Am J Respir Cell Mol Biol 2001;25:84–91. [DOI] [PubMed] [Google Scholar]
- 34.Pol A, Pfundt R, Zeeuwen P, Molhuizen H, Schalkwijk J. Transcriptional regulation of the elafin gene in human keratinocytes. J Invest Dermatol 2003;120:301–307. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Magit D, Pardee AB, Sager R. Re-expression of elafin in 21MT2 breast carcinomas by phorbol 12-myristate 13-acetate is mediated by the Ap1 site in the elafin promoter. Cancer Res 1997;57:4631–4636. [PubMed] [Google Scholar]
- 36.McKenzie CA, Abecasis GR, Keavney B, Forrester T, Ratcliffe PJ, Julier C, Connell JM, Bennett F, McFarlane-Anderson N, Lathrop GM, et al. Trans-ethnic fine mapping of a quantitative trait locus for circulating angiotensin I-converting enzyme (ACE). Hum Mol Genet 2001;10:1077–1084. [DOI] [PubMed] [Google Scholar]
- 37.Wacholder S, Rothman N, Caporaso N. Counterpoint: bias from population stratification is not a major threat to the validity of conclusions from epidemiological studies of common polymorphisms and cancer. Cancer Epidemiol Biomarkers Prev 2002;11:513–520. [PubMed] [Google Scholar]
- 38.Price AL, Butler J, Patterson N, Capelli C, Pascali VL, Scarnicci F, Ruiz-Linares A, Groop L, Saetta AA, Korkolopoulou P, et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet 2008;4:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]