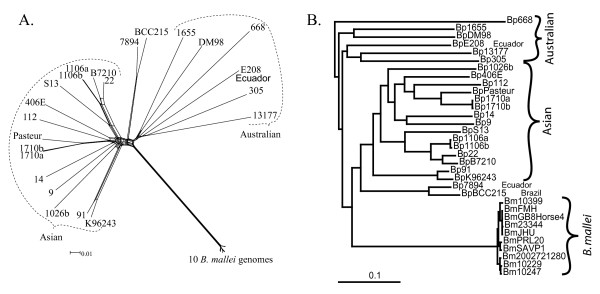
Figure 2.
Phylogenetic analysis of 14,544 single nucleotide polymorphisms shared among 33 whole genome sequences from two species. Splits decomposition analysis provides a visual account of character state conflict within the dataset (A). Bayesian phylogenetic analysis of these SNPs result in clade credibility values of 1.00 for all bifurcations (except 23344/JHU = 0.96) (B). B. thailandensis was used to root this tree (see Additional file 3).