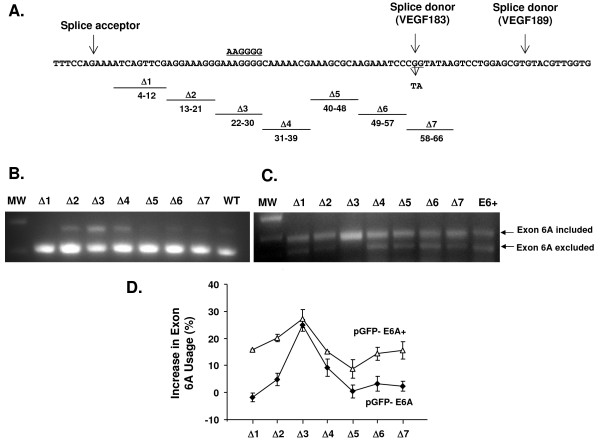
Figure 4.
Effect of deletions in exon 6A on splicing activity of pGFP-E6A minigenes. A. Sequence of full-length human VEGF exon 6A with the splice donor and acceptor sites indicated with black arrows. The naturally occurring mutation (GG mutated to TA) in exon 6A overlapping with the splice donor for VEGF183 is shown below the sequence row. Seven sequential nine nucleotide long sequential deletions were made across VEGF exon 6A in the context of pGFP-E6A and pGFP-E6A+ plasmids, excluding the sequences near splice donor and splice acceptor sites. The putative exonic splicing silencer is shown above the sequence (underlined). B. RT-PCR analysis of splicing pattern 48 hr following transfection of pGFP-E6A deletion plasmids into HEK293 cells. The anticipated positions of the products from the splicing events with inclusion or exclusion of exon 6A are indicated by arrows. C. RT-PCR analysis of splicing pattern 48 hr following transfection of pGFP-E6A+ deletion plasmids into HEK293 cell. The anticipated positions of the products for the splicing events with inclusion or exclusion of exon 6A are indicated by arrows. MW = lane with molecular weight markers. D. The percentage of exon 6A inclusion was calculated following densitometry of the gel as [E6A included band/(E6A included band + E6A excluded band)] ×100 followed by subtraction of wildtype background. Experiments were repeated 4 times in the context of pGFP-E6A (closed symbols) or three time in the context of pGFP-E6A+ (open symbols) and the average % inclusion from repeated transfections and the standard error are shown.