Figure 1.
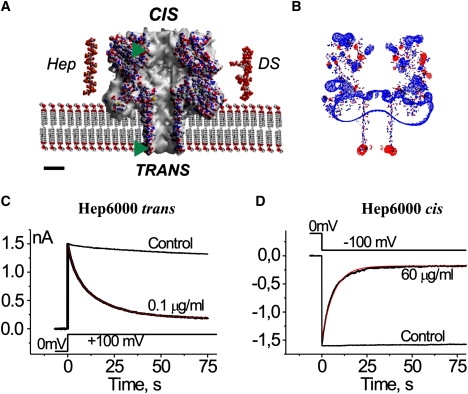
Representations of (A) the αHL channel, heparin, and DS used in this work, (B) the charge distribution at the αHL channel, and (C and D) the side-dependent heparin effect. (A) Cross section through the αHL channel (Protein Data Bank, 7AHL.pdb) embedded in membrane. Structure of the dodecameric heparin (Protein Data Bank, 1HPN.pdb; molecular mass of 3453.64 g/mol) and eventual structure of the hexadecameric DS (molecular mass of 5009.56 g/mol) were built with Chem3D (CambridgeSoft). (Abbreviations: Hep, heparin; DS, dextran sulfate.) The atoms are represented using the following color codes: O (which possesses a negative charge) in red, C in gray, H in white, N (which possesses a positive charge) in blue, and S in yellow. (Green triangles) Levels where the novel Cys and its charged derivatives are located in the channel structure. Scale bar is 2 nm. (B) The charge distribution was calculated using Coulomb calculation method (Swiss-PDBViewer, Ver. 3.7) assuming solvent ionic strength of 0.15 mol dm−3, equal to that of the 50 mM CaCl2 solution mainly used in this study. The isopotential contouring values, equal to 1.8 kT/e, are shown (red, positive potentials; blue, negative potentials). (C and D) The current decays after application of 100 mV steps to multichannel bilayers in the presence of Hep6000 at the trans (C) and cis (D) compartments of the experimental chamber. (Red lines) Best-fit of a single exponential function. Control traces (no PA) are shown for comparison. Concentrations of Hep6000, voltage protocols, current, and timescales are given in the figure. Note that the current inhibition is not total. All other conditions for the experiment are described in Materials and Methods.
