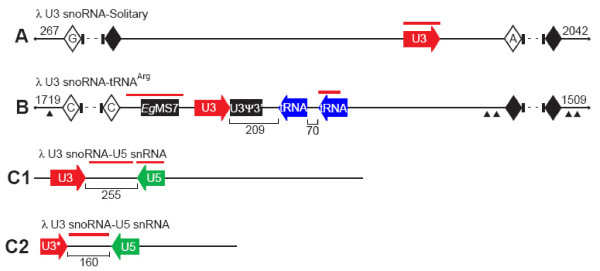
Figure 2.
Three different U3 snoRNA gene arrangements in λ clones of Euglena genomic DNA. Clone maps of (A) λ U3-Solitary (total size ~12 kbp; 4824 bp sequenced; U3-containing portion 2515 bp), (B) λ U3-tRNAArg (total size ~14 kbp; 6096 bp sequenced; U3-containing portion 2868 bp) and (C1 and C2) λ U3-U5 (total sizes 1772 and 516 bp, respectively) are shown. Terminal sequences of specified length (bp) do not contain any features of interest. Sizes of the intergenic spacers are indicated by the numbers between the various genes. Red lines demarcate the positions of probes used in Southern hybridization experiments. Repetitive elements, such as the Euglena EgMS7 microsatellite [47] (filled rectangle), the U3 snoRNA pseudo-3'-end repeat (U3Ψ3' filled rectangle) and repetitive sequences also found within certain Euglena γ-tubulin introns (filled triangles) are indicated. Solid diamonds denote simple sequence repeats that precluded effective primer design for additional primer walking. Open diamonds with letters denote homopolymer runs (A, C or G) that prematurely terminated sequencing. Due to the technical challenges posed by the many repetitive sequence elements, the λ U3-Solitary and λ U3-tRNAArg clones were not sequenced in their entirety; dashed lines denote unsequenced portions. In the λ U3-U5-C2 clone, U3* refers to a 5' truncation of the U3 gene as a result of cloning. Clone maps not drawn to scale.