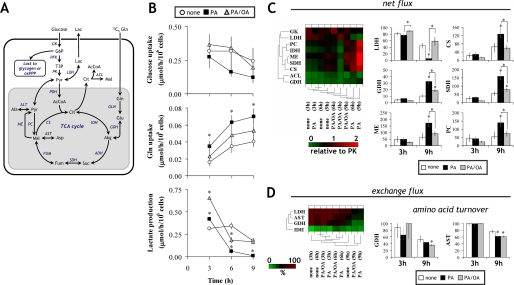
FIGURE 2.
Stable isotopic metabolic flux analysis of central carbon pathways in H4IIEC3 cells. Metabolic flux analysis of central carbon pathways using [U-13C5]glutamine at 3, 6, and 9 h. A, a metabolic network model for flux estimation in hepatoma cultures. A total of 19 net and seven exchange fluxes were calculated as described under “Experimental Procedures.” AcCoA, acetyl-CoA; Akg, α-ketoglutarate; Cit, citrate; Fum, fumarate; G6P, glucose 6-phosphate; Lac, lactate; Mal, malate; Pyr, pyruvate; Suc, succinate; T3P, triose 3-phosphate; AST, aspartate aminotransferase; ACL, ATP-citrate lyase; ADH, α-ketoglutarate dehydrogenase; CS, citrate synthase; FUM, fumarase; GDH, glutamate dehydrogenase; GLN, glutaminase; IDH, isocitrate dehydrogenase; LDH, lactate dehydrogenase; ME, malic enzyme; PDH, pyruvate dehydrogenase; PGI, phosphoglucose isomerase; SDH, succinate dehydrogenase. B, external fluxes of glucose uptake, glutamine uptake, and lactate production calculated from extracellular glucose, glutamine, and lactate measurements, respectively. Values are expressed as μmol/h/106 cells based on measurements over the 3-h intervals 0–3, 3–6, and 6–9 h. C and D, hierarchical clustering analysis of net (C, left) and exchange fluxes (D, left) under different FFA treatments. All net flux values are normalized to the PK flux. The net and exchange fluxes are represented by the color range from 0 (intense green) to 2 (intense red) in C and from 0 (green) to 100% (red) in D, respectively. Representative fluxes are shown on the right-hand sides of C and D. C (right), net fluxes expressed relative to a PK flux of 100. D (right), amino acid turnover. The first plot represents the extent of equilibration between Glu and mitochondrial Akg and the second between Asp and Mal. The first is computed from the GDH exchange flux, whereas the second is calculated from the Asp dilution flux. *, p < 0.05 for the comparison with untreated cells (white bars) or as indicated.