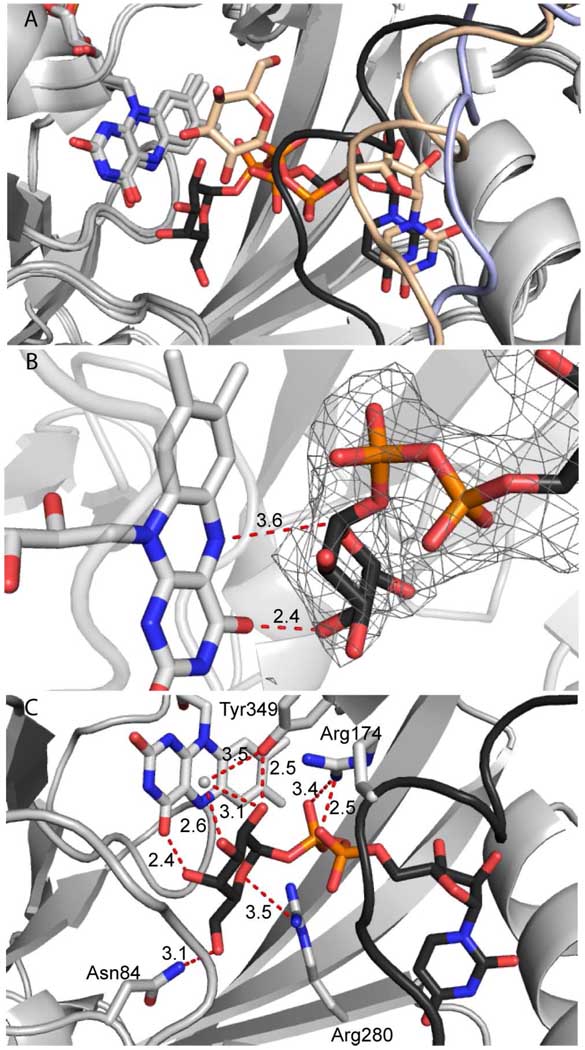
Figure 2.
Substrate binding and reduction create catalytically competent UGM active site. (A) Binding of UDP-Galp (wheat carbons) to oxidized UGM causes the mobile loop (light blue, from apo UGM pdb 2BI7 (14)) to close and form a helix (wheat). Reduction causes UDP-Galp (black carbons) and the loop (black) to shift closer to the flavin. (B) Nucleophilic flavin N5 and flavin C4 carbonyl approach the anomeric carbon and C4-OH, respectively, of the substrate UDP-Galp (distances in Å) in the reduced structure. The FO-FC omit map, calculated without ligand, is contoured two standard deviations above the mean (grey mesh). (C) Conserved residues as well as flavin and an ordered water molecule (grey sphere) orient the galactopyranose.