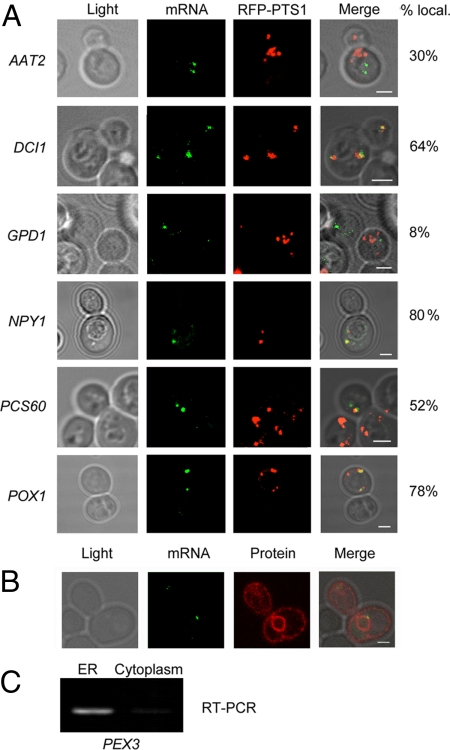
Fig. 2.
Localization of endogenous mRNAs encoding matrix proteins. (A) Representative fluorescence microscopy images of cells bearing the MS2L sequence integrated into different genes (as indicated; see the ORFINTstrains in Table S2) and transformed with plasmids expressing MS2-CP-GFP(x3) and RFP-PTS1. Cells were grown on medium containing oleate and induced with the same medium lacking methionine for 1 h before visualization. The percentage of GFP-labeled RNA granules that colocalize with peroxisomes is shown. mRNA indicates labeled RNA granules; RFP-PTS1 indicates peroxisomes. (Scale bar: 2 μm). (B) PEX3 mRNA localizes to the ER. Representative fluorescence microscopy images of MS2L-tagged PEX3 cells transformed with plasmids expressing MS2-CP-GFP(x3) and Sec63-RFP (an ER marker). mRNA indicates labeled RNA granules; protein indicates ER labeled with Sec63-RFP. The percentage of GFP-labeled granules that colocalize with ER is shown. (C) Subcellular fractionation. Yeast expressing Sec63-GFP was fractionated by density gradient centrifugation into ER and cytoplasmic fractions (same fractions as shown in fig. 9b in ref. 7). RT-PCR was performed on DNase-treated RNA derived from these fractions using PEX3-specific primers. PCR samples were then electrophoresed and visualized on a 1% agarose gel. (See fig. 9b in ref. 7 for other RNAs detected using the same conditions.)