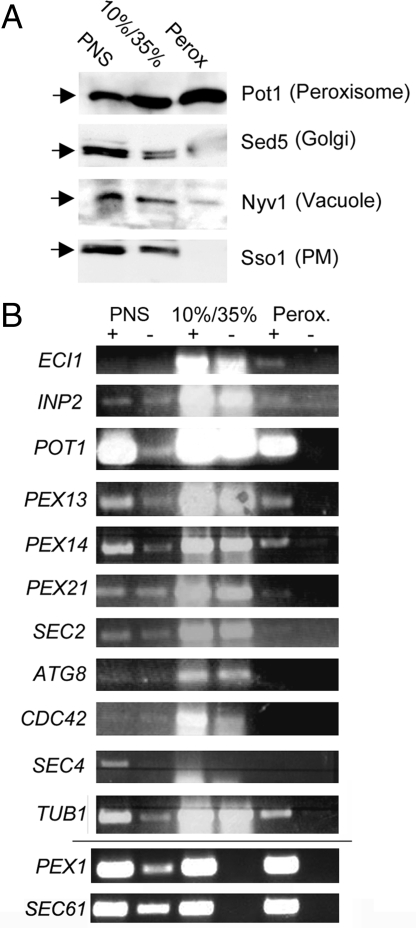
Fig. 3.
Examination of mPP localization using cell fractionation and RT-PCR. (A) Western blot analysis of peroxisome purification. Peroxisomes were purified using density gradient centrifugation followed by affinity purification. Samples (40 μg) from the 3 fractions (PNS, postnuclear supernatant; 10%/35%, mitochondria-enriched membrane fraction; Perox, purified peroxisomes) were electrophoresed on SDS/PAGE gels and analyzed by immunoblotting. Antibodies (1:5,000) against Pot1 (peroxisome), Sed5 (Golgi), Nyv1 (Vacuole), and Sso1 (PM) were used for the chemiluminescent detection of proteins. (B) RT-PCR analysis of purified peroxisomes. Yeast cells were fractionated, and samples from the PNS, 10%/35%, and peroxisomal fractions were collected, from which RNA was purified. After DNase treatment and reverse transcription (RT; +), PCR was performed using gene-specific primers (as indicated). Samples without RT (-) were used as controls for DNA contamination. After PCR, samples were electrophoresed on agarose gels and documented. PEX1 and SEC61 mRNAs were detected in a parallel experiment.