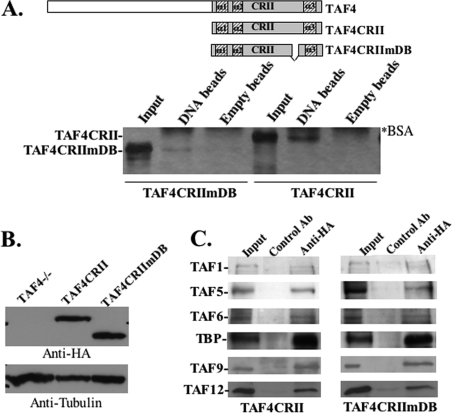
FIGURE 4.
TAF4 DNA binding is not required for TAF4-mediated growth suppression. In A: top panel, schematic representation TAF4CRII TAF4CRIImDB relative to the full-length TAF4. The mutation in TAF4CRIImDB corresponds to amino acids 1011–1051 of the spacer domain. Lower panel, TAF4CRII and TAF4CRIImDB were fused to glutathione S-transferase, expressed in E. coli, and analyzed for binding to DNA-cellulose beads (DNA lanes). Binding to empty cellulose beads (Empty beads lanes) served as a control. The input represents 10% of the protein used for binding. 20% of the eluted proteins were analyzed by SDS-PAGE and silver staining. Positions of the protein are marked on the left, and the proteins fused in binding assays are indicated at the bottom. The asterisk indicates the bovine serum albumin that is added to the binding and elution buffers. B, TAF4−/− fibroblasts were transfected with HA-TAF4CRII and HA-TAF4CRIImDB, and an empty expression vector as a control. Stable clones were analyzed by immunoblot using anti-HA and anti-tubulin monoclonal antibodies. C, total cell extracts from TAF4CRII and TAF4CRIImDB cell lines were immunoprecipitated and assayed with non-relevant control and anti-HA antibodies as indicated at the top. The immunoprecipitated complexes were then subjected to immunoblot analysis with antibodies against a subset of TAFs and TBP antibodies as indicated.