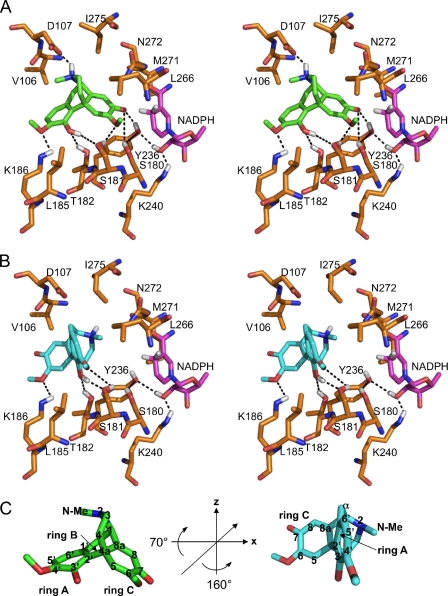
FIGURE 2.
Substrate binding pocket of SalR with docked ligand. A, stereo view of salutaridine binding in the catalytic mode. B, stereo view of salutaridine binding in the non-productive mode. C, rotation of the substrate to adopt the non-productive orientation. In C, the 3-fold symmetry axis is indicated by a line in the substrate on the left, and the rotation angles are provided in the center. The carbon numbering of salutaridine is indicated when the respective atom is visible. For A–C, the colors for the carbon skeleton of each structure are gold for amino acids, magenta for NADPH, green for salutaridine binding in the catalytic mode, or turquoise for salutaridine binding in the non-productive mode. Hydrogen, nitrogen, oxygen, and sulfur atoms are shown in white, blue, red, and yellow, respectively. Only hydrogen atoms participating in hydrogen bonding are shown. Phe104 was omitted for clarity of the stereo view, and is located behind the substrate. Putative hydrogen bonds are indicated by dashed lines. Ser180, Tyr236, and Lys240 constitute the catalytic triad. Because it has been reported that the catalytic Ser might interact with the substrate or with Tyr, both scenarios are indicated in A.