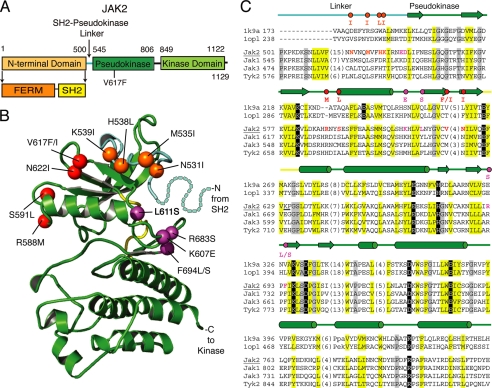
FIGURE 1.
JAK2 activating mutations are located in the pseudokinase domain and in the SH2-pseudokinase linker. A, domain organization of JAK2. B, ribbon diagram representing the JAK2 pseudokinase structure model rendered using PyMOL. The pseudokinase domain is in green, the linker region is in cyan, and the kinase hinge is in yellow. Positions of activating mutations are indicated with spheres; linker region mutations are in orange, V617F surface mutations are in red, and hinge proximal mutations are in magenta. Residues corresponding to a portion of the linker not modeled are depicted with dots. C, vertebrate JAK sequence representatives (JAK2, JAK1, JAK3, and TYK2) aligned with two kinase structure sequences (Protein Data Bank codes 1k9a and 1opl). Residue numbers are indicated to the left and right of the sequence. Common secondary structure elements are noted above the alignment and are colored according to functional domains: cyan, linker; green, pseudokinase domain; yellow, hinge. Residues within the alignment are highlighted according to conserved properties: yellow, generally hydrophobic positions; gray, generally small positions; black, polar conservations that probably contribute to function. Residues in positions of activating mutations are colored as above, with the activating mutations indicated above the alignment. Hinge region residues are underlined. The JAK2 sequence used to build the model structure is underlined.