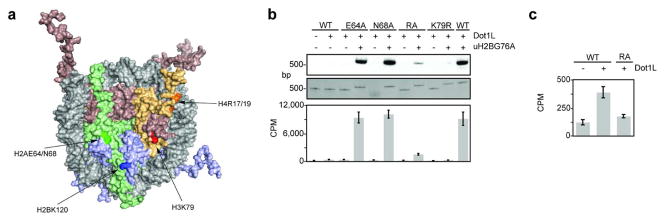
Figure 5. Dot1L assays on mutant nucleosomes.
a, Surface representation of nucleosome structure (1KX5) (49) with H2A, H2B, H3, and H4 shown in green, blue, red, and orange, respectively. Amino acids in each histone are emphasized by darker shades. b, Dot1L methyltransferase assay on mutant nucleosomes containing H2B or uH2BG76A. Nucleosomes methylated with 3H SAM were separated on native gels and stained with ethidium bromide (middle panel) prior to probing for 3H methyl incorporation by fluorography (top panel). Quantification of methyltransferase activity was performed by filter-binding followed by liquid scintillation counting (bottom panel). Error bars represent one s. d. (n = 3). c, Representation of liquid scintillation counting data in c, illustrating the effect of the H4R17/19A on Dot1L-mediated methylation of unmodified nucleosomes