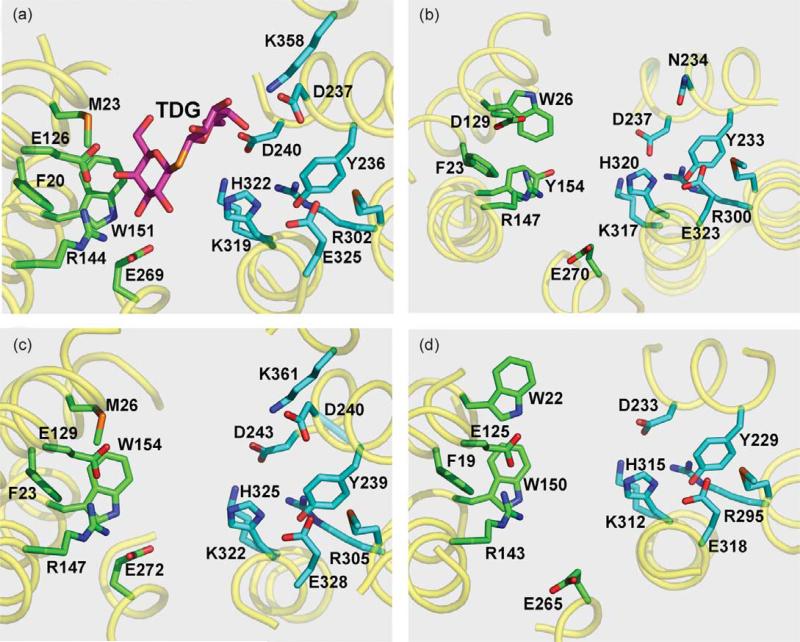
Figure 2.
Cytoplasmic view of substrate binding and H+ translocation sites of LacY and homologs. Amino acid residues known (a), or predicted ((b)–(d)) to be involved in sugar binding (green) or H+ translocation (cyan) are shown as sticks. Essential residues are numbered according to protein sequences deposited in SwissProt Databank. (a) X-ray structure of LacY (PDB code 1PV7) with bound sugar β-d-galactopyranosyl-1-thio-β-d-galactopyranoside (TDG); (b) E. coli sucrose permease CscB; (c) E. coli raffinose permease RafB; (d) B. megaterium fructose permease FruP.