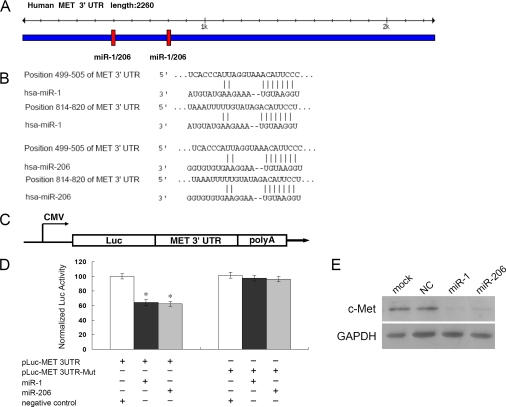
FIGURE 4.
c-Met is a direct target of miR-1/206. A, predicted miR-1/206 binding sites in c-Met 3′-UTR. Specific locations of the binding sites were marked with red color, and c-Met 3′-UTR was marked with blue color. B, alignment between the predicted miR-1/206 target sites and miR-1/206 is shown. The conserved, 7-bp seed sequence for miR-1/206:mRNA pairing is also indicated. C, diagram depicting the pMIR luciferase reporter constructs, containing a cytomegalovirus (CMV) promoter, which was utilized to verify the putative miR-1/206 binding sites (see “Experimental Procedures”). Luc, luciferase; poly A, poly(A) tail. D, HEK293 cells were co-transfected with miR-1 or miR-206, pLuc-MET 3′-UTR, along with a pRL-SV40 reporter plasmid. After 24 h, the luciferase activity was measured. Values are presented as relative luciferase activity after normalization to Renilla luciferase activity. The data are expressed as the mean value ± S.E. (error bars) of the results obtained from three independent experiments. *, differences in luciferase activity between miR-1/206 and negative control transfected cells were significant, p < 0.01. E, c-Met expression levels in RD cells after transfection with miR-1/206 were determined by Western blot analysis. As compared with the NC miRNA, miR-1/206 expression dramatically reduced the levels of c-Met in RD cells. GAPDH was used as an internal control.