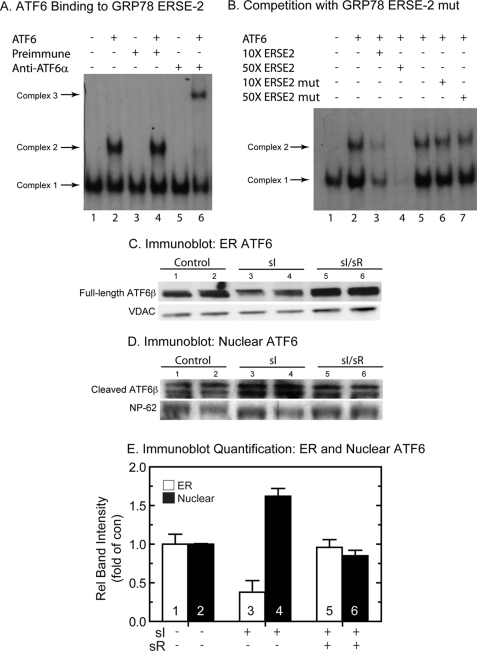
FIGURE 5.
Electromobility shift assays and ATF6 immunoblots. A, to examine ATF6 binding to the GRP78 ERSE 2, EMSA analysis was carried out as described under “Materials and Methods.” Recombinant ATF6-(116–373), used in the reactions analyzed in lanes 2, 4, and 6, was prepared by in vitro transcription/translation and then added to neonatal rat ventricular myocyte nuclear extracts, as previously described (28). The 32P-labeled GRP78 ERSE 2 probe was added to initiate the binding reactions. Complex 1 is due to direct binding of nuclear extract-derived proteins (e.g. NF-Y and YY1) to the ERSE and has been shown to be required before ATF6 will bind under these conditions. For the supershift, EMSA was carried out as described, except for the addition of either preimmune or ATF6 antiserum to lanes 3, 4, 5, and 6, as shown. B, competition binding EMSA was carried out as described above, except for the addition of 10× or 50× unlabeled GRP78 ERSE 2 oligonucleotide to lanes 3 and 4 or 10× and 50× mutated GRP78 ERSE 2 oligonucleotide to lanes 6 and 7. C and D, cultured cardiac myocytes were treated with sI or sI/R, extracted, and subjected to subcellular fractionation, as described under “Materials and Methods” (n = 2 cultures/treatment; ∼20 × 106 cells/culture). The ER fraction (C) and the nuclear fraction (D) were then analyzed by SDS-PAGE and immunoblotting for ATF6β. The region of the gel that includes the full-length endogenous ATF6 is shown in C, whereas the region of the gel that includes the cleaved active form of ATF6β is shown in D. Also shown are VDAC and NP-62, which are used as loading controls for ER and nuclear fractions, respectively. The reason for the ATF6β doublet is not known; however, it is possible that this could represent variable cleavage by S1P and/or S2P. E, immunoblots shown in C and D were quantified by densitometry. Shown are ATF6β/VDAC (ER) or ATF6β/NP-62 (Nuclear) as relative band intensity. ER and nuclear sI and sI/R values were normalized to ER and nuclear control values, respectively. n = 2 cultures/treatment, as described in C and D.