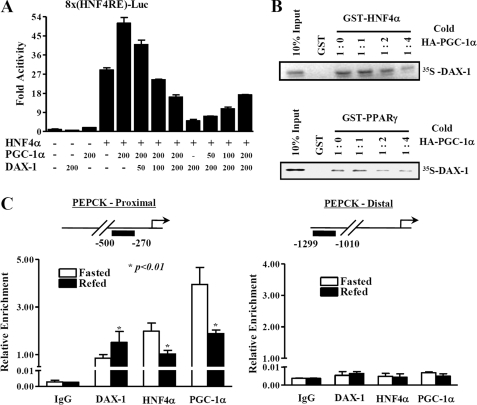
FIGURE 4.
DAX-1 competes with PGC-1α for HN4α transactivation. A, 293T cells were transfected using 200 ng of 8×(HNF4α)-Luc with the indicated amount of hnf4α, Pgc-1α, and DAX-1. Cells were harvested 40 h after transfection, and lysates were utilized for luciferase and β-galactosidase assay. The results shown are means of the β-galactosidase value from three independent experiments. Effects of DAX-1 and PGC1α alone on the basal reporter activity were also shown. B, in vitro GST competition assay was performed using in vitro translated labeled DAX-1 and unlabeled PGC-1α with bacterially expressed GST-Hnf4α (top) or with control GST-PPARγ (bottom). The ratios of labeled DAX-1 and unlabeled PGC-1α are indicated. C, quantitative analysis of the relative amounts of DAX-1, PGC-1α, and HNF4α on Pepck promoter regions using ChIP real time PCR. Recruitment of Dax-1 on the Pepck promoter is inversely correlated with the recruitment of PGC-1α and HNF4α under fasting and refeeding conditions. ChIP assays were performed using fasted (n = 4) and refed (n = 4) mouse liver samples. Liver extracts from fasted (12 h) and refed (12 h) mouse livers were chromatin-immunoprecipitated using DAX-1, HNF4α, and PGC-1α antibodies, and purified DNA samples were quantified using real time PCR using primers encompassing the proximal (lower left) and nonspecific distal (lower right) regions of the Pepck promoter (*, p < 0.01; n = 4). The upper panels show the schematic representation of the proximal (upper left) and distal (upper right) regions of the Pepck promoter and the primers used for the in vivo ChIP assay. Promoter occupancies of the indicated proteins were quantified compared with input. Data are expressed as relative enrichment of indicated proteins and Pepck promoter regions relative to that in input chromatin.