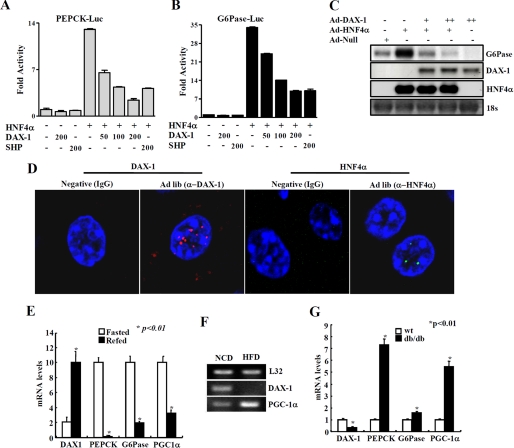
FIGURE 5.
DAX-1 inhibits HNF4α-mediated target genes and regulation of DAX-1 gene expression under different nutritional conditions. DAX-1 inhibits HNF4α target gene promoter activity and gene expression. A and B, HepG2 cells were transfected with Pepck-Luc (A) and G6Pase-Luc (B) independently with hnf4α, DAX-1, and SHP, and cell lysates were utilized for luciferase and β-galactosidase assays. Effects of DAX-1 and PGC-1α alone on the basal reporter activity were also shown. The results shown are the mean of the β-galactosidase value from three independent experiments. C, Northern blot analysis using adenovirus null, HNF4α, and DAX-1. Adenovirus for HNF4α was infected alone or with different combinations of adenovirus for DAX-1 in HepG2 cells. Then total RNA was isolated and utilized for Northern blot analysis. The same blot was hybridized with labeled G6Pase, DAX-1, and hnf4α probes. D, paraffin sections of normal (ad libitum; Ad lib) mouse liver sample were used for immunofluorescent staining. The hepatic DAX-1 and HNF4α proteins were detected with anti-DAX-1 and HNF4α antibodies and visualized with red fluorescence for DAX-1 and green fluorescence for HNF4α. Pictures are shown at ×400 magnification with a confocal microscope. DAX-1 expression under different nutritional conditions. E, quantitative PCR analysis of hepatic mRNA levels for DAX-1, PGC-1α, PEPCK, and G6Pase from fasted (24 h) or refed (24 h) mice (*, p < 0.01; n = 3). F, RT-PCR analysis of hepatic DAX-1 and PGC-1α from mice under normal chow diet or high fat diet for 8 weeks (n = 3 each). G, quantitative PCR analysis of hepatic mRNA levels for DAX-1, PGC-1α, PEPCK, and G6Pase from WT or db/db mice (*, p < 0.01; n = 3). F and G, data in E, G, and F are represented as mean ± S.D.