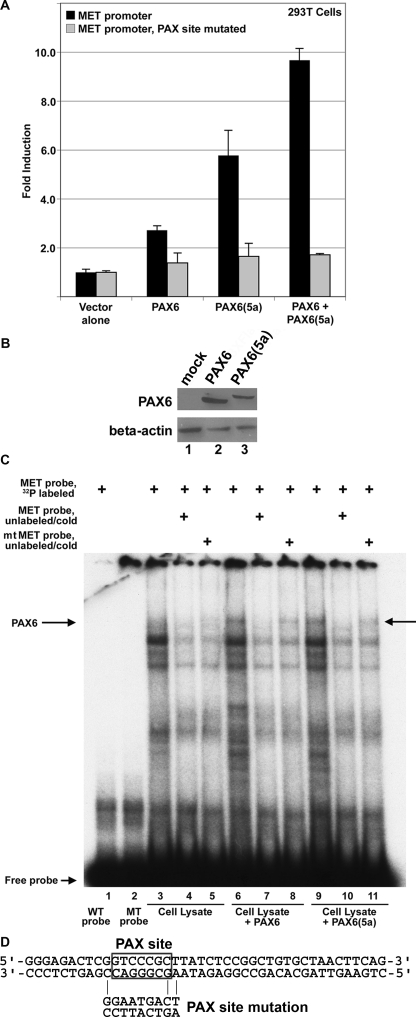
FIGURE 5.
PAX6 and PAX6(5a) activate the proximal MET promoter. A, transfection of PAX6-non-expressing 293T cells with MET promoter reporter constructs, wild type (black bars) or the paired site mutated (gray bars), with expression constructs for PAX6 and/or PAX6(5a). Total DNA transfected is equal for each sample, and the set contained both PAX6 and PAX6(5a) expression construct DNA at a 1:1 ratio. Fold induction is calculated as luciferase activity (measured in arbitrary light units) of each sample (with PAX6 and/or PAX6(5a) added) divided by luciferase levels of the reporter constructs alone. PAX6 proteins activate the MET promoter 2.7 ± 0.2-fold (canonical PAX6), 5.8 ± 0.9-fold (PAX6(5a)), or 9.7 ± 0.3-fold (both). Each bar represents n = 6. B, Western analysis for PAX6 expression in transfected 293T cells. These cells are utilized for the luciferase experiments shown in A. Cells transfected with empty vector (lane 1), a triple-FLAG-tagged PAX6 (lane 2), and a triple-FLAG-tagged PAX6(5a) (lane 3) are tested for expression of PAX6 and β-actin control. C, PAX6 and PAX6(5a) bind to the MET enhancer as shown by an EMSA. A slow migrating band (arrow labeled PAX6) is present when a radioactive probe containing the MET enhancer is combined with PAX6 protein (lane 6) or PAX6(5a) protein (lane 9). This band is absent when WT (lane 1) or mutant (MT) probe is used alone or when mock-transfected lysate is utilized (lanes 3–5). Competition with unlabeled wild type probe (lanes 4, 7, and 10) competes away PAX6/PAX6(5a) binding (lanes 7 and 10). Competition with unlabeled mutant probe (lanes 5, 8, and 11) is not as efficient in competing away protein binding to the radiolabeled probe (lanes 8 and 11), suggesting binding specificity. D, the sequence of the wild type probe used in the EMSA, with the PAX site boxed. The mutant probe is the identical sequence, except with the PAX site mutated, as shown.