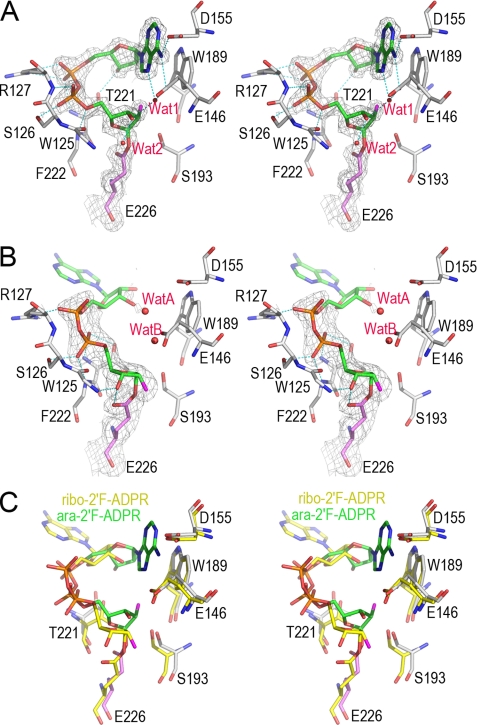
FIGURE 5.
The structural features of hydrolyzation reaction in CD38 revealed from two covalent reaction intermediates. A, the active site structure of the WTCD38·ara-2′F-ADPR complex. The active site residues are shown as sticks. Covalent adduct ara-2′F-ADPR is also shown as sticks with its carbon atoms in green. Weighted 2Fo − Fc omit electron density is shown as gray isomesh contoured at 1.2 σ covering both ara-2′F-ADPR and Glu-226. Two water molecules trapped in the active site are shown as red spheres. After cleavage and removal of nicotinamide from substrate ara-2′F-NAD, the remaining part forms a covalent adduct with the catalytic residue Glu-226, resulting in inhibition of the enzyme. After the release of the nicotinamide moiety, the adenine ring terminus can partially enter the active site, but it cannot cyclize with the terminal ribose because of the stability of the covalent intermediate and the incorrect orientation of the adenine ring, with the N1-cyclizing site pointing away. Two water molecules are present but cannot hydrolyze the intermediate, indicating its stability. B, the active site structure of the WTCD38·ribo-2′F-ADPR complex. The drawing and color scheme are the same as in A. C, structural comparison of ara-2′F-ADPR (green carbon) and ribo-2′F-ADPR (yellow carbon) complexed with CD38. The alignment shows that after the release of the nicotinamide, the adenine moiety is not able to either enter the catalytic site or enter in a wrong orientation. Both conformations of the adenine moiety favor the hydrolyzation reaction to take place.