Abstract
All mammalian cells depend on polyamines for normal growth and proliferation, but the exact roles of polyamines at the molecular level remain largely unknown. The RNA-binding protein HuR modulates the stability and translation of many target mRNAs. Here, we show that in rat intestinal epithelial cells (IECs), polyamines enhanced HuR association with the 3′-untranslated region of the c-Myc mRNA by increasing HuR phosphorylation by Chk2, in turn promoting c-Myc translation. Depletion of cellular polyamines inhibited Chk2 and reduced the affinity of HuR for c-Myc mRNA; these effects were completely reversed by addition of the polyamine putrescine or by Chk2 overexpression. In cells with high content of cellular polyamines, HuR silencing or Chk2 silencing reduced c-Myc translation and c-Myc expression levels. Our findings demonstrate that polyamines regulate c-Myc translation in IECs through HuR phosphorylation by Chk2 and provide new insight into the molecular functions of cellular polyamines.
INTRODUCTION
Polyamines (putrescine, spermidine, and spermine) are ubiquitous polycationic molecules that are found in all eukaryotic cells and are implicated in many aspects of cellular physiology (Gerner and Meyskens, 2004; Casero et al., 2007; Seiler and Raul, 2007). The intracellular polyamine content is tightly regulated by the proliferative status and depends on the dynamic balance among polyamine biosynthesis, degradation, and transport (Wang and Johnson, 1991; Gerner and Meyskens, 2004). Because of the absolute requirement for polyamines for cell growth and proliferation, the polyamine metabolic pathway has been an attractive target for antineoplastic intervention (Casero et al., 2007). The levels of cellular polyamines increase rapidly in cells stimulated to grow and divide (Belting et al., 2002; Paasinen-Sohns et al., 2000), whereas decreasing cellular polyamines stops cell cycle progression, resulting in arrest in the G1 phase (Wang, 2007). Studies from our laboratory and others (Li et al., 1999, 2002; McCormack and Johnson, 1991; Wang et al., 1991, 1993; Ray et al., 1999; Seiler and Raul, 2007; Xiao et al., 2007; Zhang et al., 2007) showed that in normal intestinal mucosa, growth and repair after injury are dependent on the supply of polyamines to the dividing cells in the crypts and that reductions in cellular polyamines by inhibiting ornithine decarboxylase (ODC), the first rate-limiting enzyme in polyamine biosynthesis, represses intestinal epithelial cell (IEC) renewal and delays wound healing in vivo and in vitro. Although the precise molecular processes governed by polyamines are not fully understood, polyamines are shown to regulate intestinal epithelial integrity by modulating the expression of growth-related genes (Patel and Wang, 1999; Liu et al., 2006).
Besides regulating transcription, polyamines also potently modulate gene expression posttranscriptionally (Wang, 2007). For example, increased levels of cellular polyamines by ectopic ODC overexpression inhibit the expression of growth-inhibitory genes such as p53 (Kramer et al., 2001; Li et al., 2001); nucleophosmin (NPM) (Zou et al., 2005, 2006); JunD and activating transcription factor-2 (ATF2) (Xiao et al., 2007); and transforming growth factor-β (Patel et al., 1998) by increasing the degradation of their mRNAs, thus contributing to the stimulation of IEC proliferation, whereas polyamine depletion increases these protein levels through stabilization of their gene transcripts, leading to growth arrest. Posttranscriptional gene regulation, which includes processes such as mRNA transport, turnover, and translation, involves specific mRNA sequences (cis-element) that interact with trans-acting factors such as RNA-binding proteins (RBPs) and microRNAs (Wilusz and Wiluszm, 2004; Keene, 2007). U- and AU-rich elements (AREs) are the best-characterized cis-acting sequences located in the 3′-untranslated regions (3′-UTRs) of many labile mRNAs (Chen and Shyu, 1995). Although AREs often function as decay elements (Bakheet et al., 2001; Wilusz et al., 2001), they also regulate translation and mRNA export (Espel, 2005). Several RBPs, including AUF1, BRF1, TTP, and KSRP, have been shown to promote ARE–mRNA decay through the recruitment of the ARE-bearing mRNA to sites of mRNA degradation such as the exosome, the proteasome, or processing bodies (Carballo et al., 1998; Laroia et al., 1999; Gherzi et al., 2004; Kedersha et al., 2005). RBPs that stabilize target mRNAs and stimulate translation include the Hu/ELAV proteins, which comprise a family of three primary neuronal members (HuB, HuC, and HuD) and one ubiquitous member HuR (Gorospe,, 2003; Hinman and Lou, 2008).
HuR is characterized by the presence of two N-terminal RNA recognition motifs (RRMs) with high affinity for AREs, followed by a nucleocytoplasmic shuttling sequence and a C-terminal RRM that recognizes the poly(A) tail (Fan and Steitz, 1998) and has emerged as a key regulator of genes that are central to cell proliferation, stress response, immune cell activation, carcinogenesis, and replicative senescence (López de Silanes et al., 2004a; Hinman and Lou, 2008). HuR is predominantly nuclear in unstimulated cells, but it rapidly translocates to the cytoplasm, where it stabilizes specific mRNAs and affects the translation of several target mRNAs, repressing translation in some instances (as shown for p27, Wnt5a, and type 1 insulin-like growth factor receptor), activating translation in other instances (as shown for prothymosin α, p53, mitogen-activated protein kinase phosphatase-1, hypoxia inducible factor-1α, and cytochrome c; Mazan-Mamczarz et al., 2003; reviewed in Abdelmohsen et al., 2008). However, HuR is constitutively expressed and an RNA signature motif recognized by HuR is widely found in numerous ARE- and non–ARE-containing mRNAs (López de Silanes et al., 2004b), suggesting that HuR can bind to a broad range of mRNAs and can influence their posttranscriptional fates. Additional layers of HuR regulation of target mRNAs have been uncovered in the past few years. It was recently shown that the checkpoint kinase Chk2 phosphorylates HuR and thereby altered the affinity of HuR for its target transcripts after exposure to oxidative stress (Abdelmohsen et al., 2007). In addition, HuR phosphorylation by protein kinase Cα elevates its cytoplasmic abundance (Doller et al., 2007), whereas cyclin-dependent kinase-1–mediated HuR phosphorylation prevents the cytoplasmic accumulation of HuR (Kim, 2008). We recently demonstrated that polyamines also modulate the subcellular distribution of HuR through AMP-activated protein kinase and that polyamine depletion increases the cytoplasmic levels of HuR (Zou et al., 2008).
The c-Myc gene encodes a nuclear transcription factor that plays an important role in the regulation of cell proliferation, differentiation, apoptosis, and the cell cycle (Ryan and Birnie, 1996; Trumpp et al., 2001). c-Myc expression is controlled at multiple levels, including transcription (Marcu et al., 1992), stability of both the mRNA and protein (Ross, 1995), and translation (Nanbru et al., 1997; Kim et al., 2003; Liao et al., 2007; Wall et al., 2008). Although c-Myc transcription requires polyamines (Celano et al., 1989; Patel and Wang, 1997), the exact mechanisms whereby cellular polyamines regulate c-Myc translation remain unknown. There have been reports indicating that the 3′-UTR of c-Myc mRNA contains AREs that interact with RBPs and are involved in the control of c-Myc translation (Lafon et al., 1998). Given our long-standing interest in understanding polyamine function in mammalian models of gut mucosal growth and repair, we examined the effects of polyamines on c-Myc translation in IECs. The results presented herein indicate that elevating the cellular levels of polyamines enhanced HuR association with c-Myc mRNA through Chk2-regulated HuR phosphorylation, promoted c-Myc translation, and contributed to an elevation in c-Myc steady-state levels. In contrast, polyamine depletion in IECs decreased [HuR/c-Myc mRNA] complexes by repressing HuR phosphorylation, in turn inhibiting c-Myc translation.
MATERIALS AND METHODS
Cell Culture and Supplies
IEC-6 cells, derived from normal rat intestinal crypt cells, are nontumorigenic and retain the undifferentiated characteristics of intestinal crypt cells (Quaroni et al., 1979). ODC-overexpressing IEC-6 (ODC-IEC) cells were developed from IEC-6 cells as described previously (Liu et al., 2006) and expressed a more stable ODC variant with full enzyme activity (Ghoda et al., 1989). IEC-6 cells, ODC-IEC cells, and Caco-2 cells were maintained in DMEM supplemented with 5% heat-inactivated fetal bovine serum and antibiotics. The anti-HuR antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the antibody against all phosphorylated proteins was obtained from Zymed Laboratories (South San Francisco, CA).
Plasmid Construction
The Chk2 expression vector was described previously (Abdelmohsen et al., 2007). The vectors expressing wild-type HuR-TAP (tandem affinity purification) fusion proteins or point-mutated HuR-TAP fusion proteins were generated by site-directed mutagenesis as described previously (Abdelmohsen et al., 2007). The chimeric firefly luciferase reporter construct containing the c-Myc 3′-UTR was generated as described previously (Liao et al., 2007). The 456-base pair ARE fragment from the c-Myc 3′-UTR was amplified and subcloned into the pGL3-Luc plasmid (Promega, Madison, WI) to generate the chimeric pGL3-Luc-c-Myc-3′-UTR. Luciferase activity was measured using the Dual Luciferase Assay System (Promega) following the manufacturer's instructions. The firefly-to-Renilla luciferase activity ratio was further compared with the levels of each luciferase mRNA.
Western Blot Analysis
Whole-cell lysates were prepared using 2% SDS, sonicated, and centrifuged at 4°C for 15 min. The supernatants were boiled for 5 min and size-fractionated by SDS-polyacrylamide gel electrophoresis (PAGE). After transferring proteins onto nitrocellulose filters, the blots were incubated with primary antibodies recognizing c-Myc, HuR, and Chk2 proteins; after incubations with secondary antibodies, immunocomplexes were developed by using chemiluminescence.
Reverse Transcription (RT) followed by Polymerase Chain Reaction (PCR) and Real-Time Quantitative (q)PCR Analysis
Total RNA was isolated from cells after different treatments by using RNeasy mini kit (QIAGEN, Valencia, CA) and used in reverse transcription and PCR amplification reactions as described previously (Zou et al., 2006). The levels of β-actin PCR product were assessed to monitor the even RNA input in RT-qPCR samples. RT-qPCR was performed using 7500-Fast Real-Time PCR Systems (Applied Biosystems, Foster City, CA) with specific primers, probes, and software (Applied Biosystems).
Analysis of Newly Translated Protein and Polysome Analysis
New synthesis of c-Myc protein was measured by l-[35S]methionine and l-[35S]cysteine incorporation assays as described previously (Kim, 2008). Cells were incubated with 1 mCi (1 Ci = 37 GBq) of l-[35S]methionine and l-[35S]cysteine per 60-mm plate for 20 min, whereupon cells were lysed using radioimmunoprecipitation assay buffer. Immunoprecipitations were carried out for 1 h at 4°C by using either a polyclonal antibody recognizing c-Myc or immunoglobulin G (IgG)1 (BD Biosciences Pharmingen, San Diego, CA). After extensive washes in TNN buffer (50 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM EDTA, and 0.5% NP-40), the immunoprecipitated material was resolved by 10% SDS-PAGE, transferred onto polyvinylidene difluoride filters, and visualized with a PhosphorImager (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom).
Polysome analysis was performed as described previously (Kawai et al., 2006). In brief, IEC-6 cells at ∼70% confluence were incubated for 15 min in 0.1 mg/ml cycloheximide and then lifted by scraping in 1 ml of PEB lysis buffer (0.3 M NaCl, 15 mM MgCl2, 15 mM Tris-HCl, pH 7.6, 1% Triton X-100, 1 mg/ml heparin, and 0.1 mg/ml cycloheximide) and lysed on ice for 10 min. Nuclei were pelleted (10,000 × g; 10 min), and the resulting supernatant was fractionated through a 10–50% linear sucrose gradient to fractionate cytoplasmic components according to their molecular weights. The eluted fractions were prepared with a fraction collector (Brandel, Gaithersburg, MD), and their quality was monitored at 254 nm by using a UV-6 detector (ISCO, Lincoln, NE). After RNA in each fraction was extracted with 8 M guanidine-HCl, the levels of each individual mRNA were quantified by RT-qPCR in each of the fractions, and their abundance represented as a percent of the total mRNA in the gradient.
Binding Assays: Biotin Pull-Down and Ribonucleoprotein (RNP) Immunoprecipitation (IP) Analyses
The synthesis of biotinylated transcripts and analysis of RBPs bound to biotinylated RNA were done as described previously (Xiao et al., 2007) and explained in detail in Supplemental Data.
IP of endogenous RNA–protein complexes was performed as described previously (López de Silanes et al., 2004b). The RNA isolated from IP materials was reverse transcribed by using random hexamers or oligo(dT) primer and SSII Reverse Transcriptase (Invitrogen, Carlsbad, CA). Conditions for qPCR and oligomers to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ProTα products were described previously (Zou et al., 2006). Oligomers used for PCR products are listed in the Supplemental Data.
Statistics
Values are means ± SE from three to six samples. Autoradiographic and immunoblotting results were repeated three times. The significance of the difference between means was determined by analysis of variance. The level of significance was determined using Duncan's multiple range test (Harter, 1960).
RESULTS
Polyamines Regulate c-Myc mRNA Translation via Its 3′-UTR
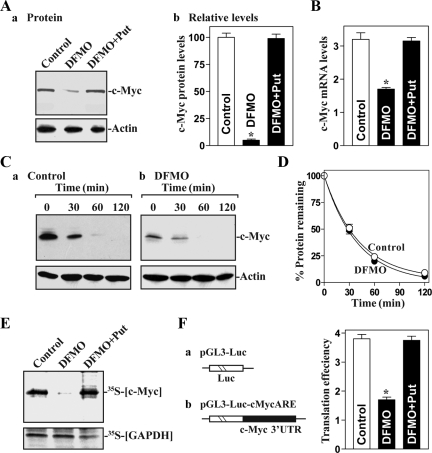
The regulation of c-Myc translation by polyamines was revealed in two sets of experiments. First, we examined the changes in levels of c-Myc mRNA and protein after the depletion of cellular polyamines. Exposure of IEC-6 cells to 5 mM dl-α-difluoromethylornithine (DFMO), a specific inhibitor of polyamine biosynthesis, for 4 d inhibited ODC enzyme activity and virtually depleted cellular polyamines, as reported previously (Liu et al., 2006). The levels of putrescine and spermidine were undetectable on day 4 after treatment with DFMO, and spermine was decreased by ∼60% (data not shown). Polyamine depletion significantly decreased the steady-state levels of both c-Myc protein and mRNA. However, quantitative analysis of these results indicated that the levels of c-Myc protein decreased by >90% in polyamine-deficient cells (Figure 1A), whereas c-Myc mRNA levels decreased only by ∼50% (Figure 1B). The specificity of these effects was demonstrated by the addition of exogenous polyamine putrescine (10 μM), which prevented the decrease in c-Myc mRNA and protein. The results presented in Figure 1, C and D, further show that c-Myc protein stability (as measured by incubating cells with cycloheximide to block de novo protein synthesis) was not affected by polyamine depletion, supporting the view that polyamines regulate c-Myc translation. To investigate directly whether decreased c-Myc translation might also contribute to reducing c-Myc expression after polyamine depletion, we compared the rate of new c-Myc synthesis between control cells and cells treated with DFMO alone or DFMO plus putrescine. Cells were incubated in the presence of l-[35S]methionine and l-[35S]cysteine for 20 min, whereupon newly translated c-Myc was visualized by IP. The brief incubation period was chosen to minimize the contribution of c-Myc degradation in our analysis. As shown in Figure 1E, newly synthesized c-Myc was markedly lower in DFMO-treated cells, whereas exogenous putrescine given together with DFMO restored the rate of new c-Myc synthesis to normal levels. Polyamine depletion did not affect global protein translation (Supplemental Figure A1), nor did it influence nascent GAPDH translation, as the rates of newly synthesized GAPDH protein in control cells were similar to those in cells treated with DFMO alone or DFMO plus putrescine (Figure 1E).
Figure 1.
Polyamine depletion inhibits c-Myc mRNA translation. (A) Levels of c-Myc protein after polyamine depletion: a, representative immunoblots of c-Myc protein and b, quantitative analysis of derived from densitometric scans of immunoblots of c-Myc as described in a. Cells were treated with DFMO (5 mM) alone or DFMO plus putrescine (Put; 10 μM) for 4 d; levels of c-Myc protein were measured by Western blot analysis. Values are the means ± SE from three separate experiments, and relative levels of c-Myc protein were corrected for protein loading as measured using densitometry of actin. *p < 0.05 compared with controls and cells treated with DFMO plus Put. (B) Levels of c-Myc mRNA as measured by RT-qPCR analysis in cells described in A. Values are the means ± SE of data from three separate experiments. *p < 0.05 compared with controls and cells exposed to DFMO plus Put. (C) Assessment of c-Myc protein stability: a, untreated control and b, cells treated with DFMO for 4 d. After treatment with cycloheximide (50 μg/ml) for the times indicated, whole-cell lysates were harvested for Western blot analysis. (D) Quantitative analysis of the immunoblotting signals in C, as measured by densitometry. Values are means ± SE of data from three separate experiments; the relative levels of c-Myc were corrected for protein loading by measuring β-actin densitometric signals. (E) Newly translated c-Myc protein in cells that were proposed as described in A. c-Myc translation was measured by incubating cells with l-[35S]methionine and l-[35S]cysteine for 20 min, followed by immunoprecipitation by using anti-c-Myc antibody, resolving immunoprecipitated samples by SDS-PAGE, and transferring for visualization of signals by using a PhosphorImager. The translation of housekeeping control GAPDH was measured similarly. (F) Changes in c-Myc translation efficiency as measured by using pGL3-Luc-MycARE reporter assays in cells described in A. Left, schematic of plasmids: control (pGL3-Luc); and chimeric firefly luciferase (Luc)-c-Myc 3′-UTR (pGL3-Luc-MycARE). Right, levels of c-Myc translation. The pGL3-Luc-MycARE or pGL3-Luc (negative control) was cotransfected with a Renilla luciferase reporter. Twenty-four hours later, firefly and Renilla luciferase activities were assayed. Luciferase values were normalized to the mRNA levels to obtain translation efficiencies and expressed as means ± SE of data from three separate experiments. *p < 0.05 compared with control and cells treated with DFMO plus Put.
To further confirm these findings and to examine whether the translational effect of polyamines was exerted through the ARE, we used a firefly luciferase reporter gene construct containing the c-Myc ARE within the 3′-UTR (pGL3-Luc-cMycARE) and negative control vector pGL3-Luc (Figure 1F, left). A plasmid expressing Renilla luciferase was also cotransfected as an internal control for normalization of firefly luciferase. To distinguish translational output from changes in mRNA turnover, the luciferase activity assays were normalized to luciferase-reporter mRNA levels to assess the translation efficiency. As shown in Figure 1F, right, polyamine depletion by DFMO treatment inhibited c-Myc translation as indicated by a decrease in c-Myc ARE luciferase reporter gene activity. The combined DFMO and putrescine treatment prevented the decrease in c-Myc translation, rendering the rate of c-Myc ARE-mediated translation similar to that observed in control cells. Spermidine (5 μM) had an effect equal to that of putrescine on levels of c-Myc mRNA translation when it was added to cultures that contained DFMO (data not shown). By contrast, no changes in luciferase activity were seen in response to polyamine depletion when testing a control construct without the c-Myc ARE (data not shown). These results indicate that decreasing the levels of cellular polyamines represses c-Myc mRNA translation through the c-Myc 3′-UTR ARE.
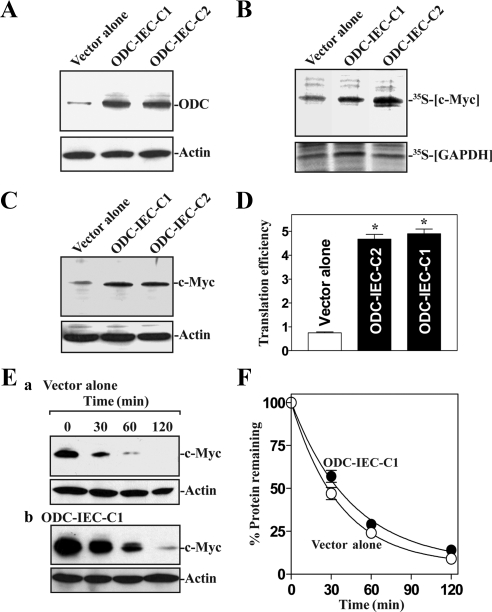
Second, we determined the effect of increasing cellular polyamines on c-Myc mRNA translation by using two clonal populations of intestinal epithelial cells stably expressing ODC (ODC-IEC) (Liu et al., 2006; Zou et al., 2006). ODC-IEC cells exhibited very high levels of ODC protein (Figure 2A) and >50-fold increase in ODC enzyme activity. Accordingly, the levels of putrescine, spermidine, and spermine in ODC-IEC cells were increased by ∼12-fold, ∼2-fold, and ∼25% compared with cells transfected with the control vector lacking ODC cDNA (data not shown), as reported previously (Liu et al., 2006). As shown in Figure 2B, newly synthesized c-Myc was significantly more abundant in stable ODC-IEC cells. Consistently, ODC-IEC cells also displayed a substantial increase in levels of c-Myc protein compared with those observed in cells transfected with the control vector (Figure 2C). This induction in c-Myc expression by increasing cellular polyamines was also partially due to an increase in c-Myc mRNA translation, because the levels in c-Myc ARE luciferase reporter gene activity were significantly increased in ODC-IEC cells (Figure 2D). The effects of ODC overexpression on c-Myc translation were not simply due to clonal variation, because two different clonal populations, ODC-IEC-C1 and ODC-IEC-C2, showed similar responses. The increase in c-Myc expression in stable ODC-IEC cells did not result from an increase in c-Myc protein stability because the half-life of c-Myc protein was not significantly different in control cells compared with cells stably overexpressing ODC (ODC-IEC cells; Figure 2, E and F). In addition, neither increasing nor decreasing polyamine levels altered global translation significantly, as assessed by measuring the rates of whole-cell protein synthesis (Supplemental Fig. A1). Together, these results indicate that increased levels of cellular polyamines enhanced c-Myc mRNA translation.
Figure 2.
Increasing cellular polyamines enhances c-Myc translation. (A) Levels of ODC protein in controls (vector alone) and stable ODC-IEC cells. IEC-6 cells were infected with either the retroviral vector containing the sequence encoding mouse ODC cDNA or control retroviral vector lacking ODC cDNA. Clones resistant to the selection medium containing 0.6 mg/ml G418 were isolated and screened for ODC expression. Levels of ODC protein were assessed by Western blot analysis. (B) Newly synthesized c-Myc protein in cells described in A. After cells were incubated with l-[35S]methionine and l-[35S]cysteine for 20 min, cell lysates were prepared and immunoprecipitated by using anti-c-Myc antibody, resolved by SDS-PAGE, and transferred for visualization of signals by using a PhosphorImager. The translation of housekeeping control GAPDH was measured similarly. (C) Representative immunoblots of c-Myc levels in cells described that were processed as described in A. (D) Changes in c-Myc translation efficiency as measured by using pGL3-Luc-MycARE reporter assays in cells described in A. The pGL3-Luc-MycARE or pGL3-Luc (negative control) was cotransfected with a Renilla luciferase reporter, and firefly and Renilla luciferase activities were assayed 24 h thereafter. Luciferase values were normalized to the mRNA levels to obtain translation efficiencies and expressed as means ± SE of data from three separate experiments. *p < 0.05 compared with the vector alone. (E) Effect of increasing cellular polyamines on c-Myc protein stability: a, vector alone and b, stable ODC-IEC cells. After incubation in the presence of cycloheximide IEC whole-cell lysates were harvested for Western blot analysis. (F) Densitometric quantitation of the Western blotting signals in E. Values are means ± SE of data from three separate experiments.
Polyamines Modulate Distribution of c-Myc mRNA in Polyribosomes
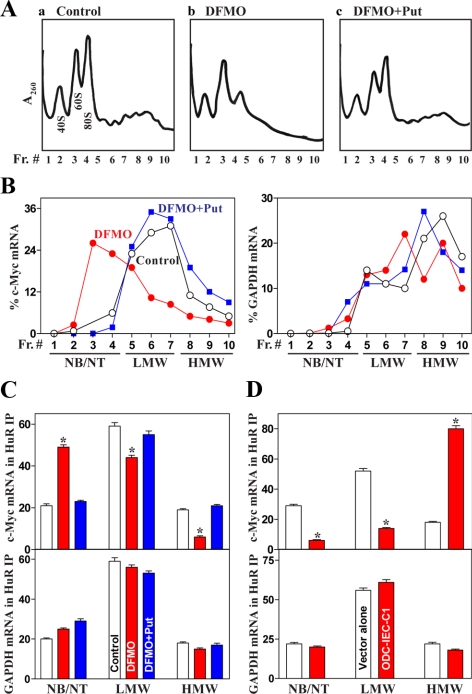
To further substantiate the effect of cellular polyamines on c-Myc mRNA translation, we first examined the relative distributions of c-Myc mRNA and its interaction with RNA-binding protein HuR on individual fractions from polyribosome gradients after polyamine depletion. HuR was chosen in this study because c-Myc mRNA contains several computationally predicted hits of the HuR signature motif and was shown to interact with HuR (Lafon et al., 1998). Polyribosome distribution profiles were examined in control cells and in cells treated with DFMO alone or DFMO plus putrescine for 4 d as described previously (Chen et al., 2008). In this study, fractions 1–4 included mRNAs that were not associated with components of the translation machinery or cosedimented with ribosome subunits (monosomes); hence, they were not considered to be translated. Fractions 5–7 included mRNAs that bound to single ribosomes or formed polysomes of low molecular weight, and they were considered to be translated at low-to-moderate levels. Fractions 8–10 included the mRNAs that were associated with polysomes of high molecular weight, and they were thus considered to be actively translated. Figure 3A showed global changes in polysomal profiles in control cells and cells treated with DFMO alone or DFMO plus putrescine. When the distribution of c-Myc mRNA levels in fractions across the gradient in control and polyamine-deficient cells was compared, it was found to be more abundant in nontranslating or low-translating fractions of polyamine-deficient cells (Figure 3B, left). Furthermore, exogenous putrescine given together with DFMO restored HuR and c-Myc mRNA polysomal distribution to that seen in untreated cells. This redistribution of c-Myc mRNA in polyribosomes after polyamine depletion was specific, as the housekeeping GAPDH mRNA distributed similarly in the three groups (Figure 3B, right). In addition, Western blot analysis of fractions across the gradient showed that HuR in control cells was abundant in fractions 1–5 but still detectable in fractions 6–8, whereas in DFMO-treated cells, HuR was predominantly located in fractions 1–3 and dramatically decreased in fraction 4 and thereafter (Supplemental Figure A2).
Figure 3.
Polysomal distributions of c-Myc mRNA and its association with HuR after modulating the levels of cellular polyamines. (A) Polysomal profiles from control cells (a) and cells exposed to DFMO alone (b) or DFMO plus Put (c) for 4 d. Nuclei were pelleted, and the resulting supernatants were fractionated through a 10-to-50% linear sucrose gradient. Three experiments were performed that showed similar results. (B) Distributions of c-Myc mRNA (left) and housekeeping GAPDH mRNA (right) in each gradient fraction prepared from all three groups. Total RNA was isolated from the different fractions, and the levels of c-Myc and GAPDH mRNAs were measured by RT-qPCR analysis and plotted as a percentage of the total c-Myc or GAPDH mRNA levels in that sample. The translational activity associated with each fraction is indicated as untranslated (NB, not bound to polysomes; NT, not translated), moderately translated (LMW, low-molecular-weight polysomes) and actively translated (HMW, high-molecular-weight polysomes). Data represent the average of three independent experiments yielding similar results. (C) Binding of HuR to c-Myc mRNA (top) and GAPDH mRNA (bottom) in various polysomal fractions in cells described in A. Fractions of NB/NT, fractions of LMW, and fractions of HMW from all three groups were used for IP in the presence of anti-HuR antibody; c-Myc and GAPDH mRNAs present in the IP materials were detected by RT-qPCR analysis. Values are means ± SE from three separate experiments. *p < 0.05 compared with control cells and cells exposed to DFMO plus Put. (D) Association of HuR to c-Myc mRNA (top) and GAPDH mRNA (bottom) in various polysomal fractions in stable ODC-expressing cells (ODC-IEC-C1). Values represent the means ± SE from three separate experiments. *p < 0.05 compared with cells transfected with the empty vector alone.
We further examined the effects of manipulating cellular polyamine levels upon the interaction of HuR with c-Myc mRNA in fractions from polysomal gradients. These interactions were tested by IP of HuR under conditions that preserved its association with target mRNAs in RNP complexes; the mRNAs present in the RNP complexes were measured by RT followed by real-time qPCR analyses. Interestingly, polyamine depletion by DFMO increased HuR association with c-Myc mRNA in nontranslating fractions but decreased it in translating fractions (Figure 3C, top). In contrast, increased levels of cellular polyamines by ODC overexpression enhanced HuR association with c-Myc mRNA in actively translating fractions (Figure 3D, top). In contrast, there were no significant changes in the association of HuR with GAPDH mRNA in different fractions across the gradients of polyamine-deficient cells and stable ODC-expressing cells compared with controls (Figure 3, C and D, bottom). These observations further supported the possibility that cellular polyamines regulate c-Myc mRNA translation through a specific process involving the RNA-binding protein HuR.
Polyamine Depletion Represses HuR Binding to the c-Myc 3′-UTR
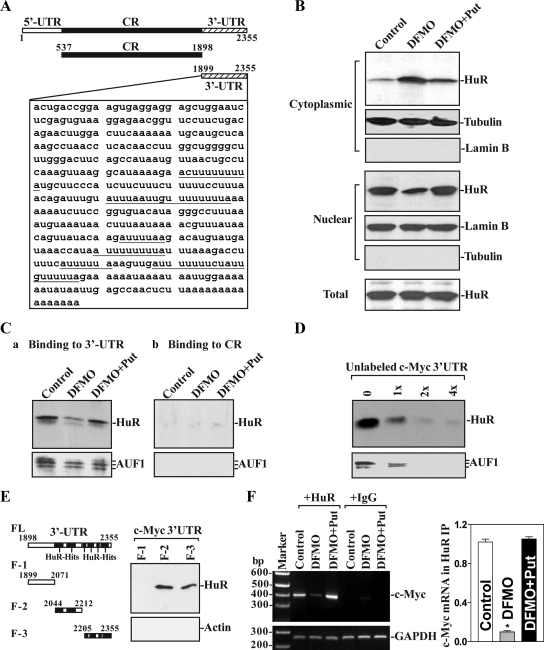
Given the predicted affinity of HuR for the 3′-UTR of the c-Myc mRNA (Figure 4A), we hypothesized that HuR bound the c-Myc 3′-UTR and further postulated that this association could be regulated by cellular polyamines. Consistent with our previous observations (Zou et al., 2006), polyamine depletion by DFMO increased cytoplasmic HuR levels and reduced its nuclear abundance but did not change its whole-cell levels (Figure 4B). Supplementation with putrescine reversed the DFMO-triggered changes in HuR subcellular distribution, as did spermidine supplementation (data not shown). To monitor the quality and abundance of the nuclear and cytoplasmic fractions, we examined the levels of lamin B (a nuclear protein) and β-tubulin (a cytoplasmic protein). Assessment of these markers revealed that there was no contamination between cytoplasmic and nuclear fractions. The change in relocalization of HuR after polyamine depletion was further confirmed by immunofluorescence analysis. Consistent with the Western blotting results and our previous findings (Zou et al., 2006), HuR immunostaining increased significantly in the cytoplasms of DFMO-treated cells, but this increase was prevented by exogenous putrescine (data not shown). These results indicate that lowering cellular polyamines increased the cytoplasmic levels of HuR.
Figure 4.
Polyamine depletion represses HuR binding to c-Myc mRNA. (A) Schematic representation of c-Myc mRNA and the HuR-hits in its 3′-UTR. (B) Representative immunoblots of HuR in different subcellular fractions isolated from control cells and cells treated with DFMO alone or DFMO plus PUT for 4 d. Whole-cell lysates, cytoplasmic, and nuclear lysates were prepared and subjected to SDS-PAGE. After detecting HuR, blots were reprobed to detect β-tubulin in cytoplasmic, lamin B in nuclear, and actin in whole-cell lysates to control for the quality of the fractionation procedure and the even loading of samples. (C) Changes in binding of HuR and AUF1 to c-Myc mRNA in cells described in B. Cytoplasmic lysates (120 μg of each) prepared from all three groups were incubated with 6 μg of biotinylated c-Myc 3′-UTR or CR for 30 min at 25°C, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. Representative immunoblots of HuR or AUF1 using the pull-down materials are shown for the 3′-UTR (a) or CR (b). (D) Effect of unlabeled c-Myc 3′-UTR added to the binding reaction; the levels of [HuR/c-Myc 3′-UTR] and [AUF1/c-Myc 3′-UTR] complexes were detected by pull-down assays. Three independent experiments were performed showing similar results. (E) HuR-binding to different fractions (F) of c-Myc 3′-UTR. Left, schematic representation of various c-Myc 3′-UTR fractions used for mapping c-Myc 3′-UTR for HuR-binding sites. Right, representative HuR immunoblots using the pull-down materials by different fractions of c-Myc 3′-UTR. Three experiments were performed that showed similar results. (F) Association of endogenous HuR with endogenous c-Myc mRNA in cells described in B. Whole-cell lysates from all three groups were used for IP in the presence of anti-HuR antibody or nonspecific IgG. Levels of c-Myc and GAPDH RNAs in the IP material were examined by RT-PCR analysis (left) and RT-qPCR analysis (right). Values are the means ± SE of data from three separate experiments. *p < 0.05 compared with control cells and cells exposed to DFMO plus Put.
To study the association of HuR with c-Myc mRNA, we used biotinylated transcripts spanning the c-Myc 3′-UTR in RNA pull-down assays using cell lysates prepared from either untreated or polyamine-deficient cells. Unexpectedly, polyamine depletion remarkably inhibited HuR association with c-Myc mRNA, even though it significantly increased cytoplasmic HuR levels (Figure 4B). As shown in Figure 4Ca, the c-Myc 3′-UTR transcript readily associated with cytoplasmic HuR, as detected by Western blot analysis of the pull-down material; the binding intensity decreased dramatically when using lysates prepared from cells that were treated with DFMO for 4 d, but it returned to normal levels when cells were treated with DFMO plus putrescine. The c-Myc 3′-UTR also formed complex with another RBP (AUF1) in IECs, but this interaction was not affected by polyamine depletion. In addition, transcripts corresponding to the coding region (CR) of c-Myc mRNA did not bind to HuR or AUF1 (Figure 4Cb). To determine the specificity of binding of the c-Myc 3′-UTR to HuR or AUF1, competition experiments were carried out. As shown in Figure 4D, the association of the c-Myc 3′-UTR with HuR or AUF1 was progressively inhibited when increasing concentrations of unlabeled c-Myc 3′-UTR were added to the binding reaction. By contrast, addition of excess unlabeled c-Myc CR did not reduce the levels of [HuR/c-Myc 3′-UTR] complexes and [AUF1/c-Myc 3′-UTR] complexes (data not shown). The results presented in Figure 4E further suggest that HuR binding to the c-Myc 3′-UTR could be mediated by up to six specific sites containing computationally predicted hits of the HuR motif. Partial biotinylated transcripts spanning the c-Myc 3′-UTR were prepared and equimolar quantities were tested for binding to HuR in pull-down assays. HuR was found to bind F-2 and F-3, two transcripts containing predicted HuR motif hits (Figure 4E, right), but not F-1, which lacks putative HuR hits.
We also examined the in vivo association of endogenous HuR with endogenous c-Myc mRNA after polyamine depletion by RNP IP assays. There was abundant c-Myc mRNA in the RNP complexes immunoprecipitated using anti-HuR antibody, as measured by conventional PCR (Figure 4F, left) and RT-qPCR analyses (Figure 4F, right). The association of endogenous c-Myc mRNA with endogenous HuR was decreased by >85% in cells treated with DFMO for 4 d but was returned to normal level when testing lysates from cells treated with putrescine and DFMO. Importantly, the c-Myc mRNA was undetectable in nonspecific IgG IPs (Figure 4F, middle). In this study, GAPDH mRNA was also examined as a negative control, because this highly abundant (housekeeping) transcript is present as a low-level contaminant in the IP materials, thus serving to monitor the equal input of lysate as reported previously (Abdelmohsen et al., 2007). Together, these findings indicate that cytoplasmic HuR specifically associates with the 3′-UTR of c-Myc mRNA and that this interaction decreases after polyamine depletion.
HuR Phosphorylation by Chk2 Is Essential for Binding to c-Myc mRNA
Although cytoplasmic HuR levels increased after polyamine depletion (Figure 4B), HuR binding to c-Myc mRNA in fact decreased significantly (Figure 4C), suggesting that posttranslational events triggered this reduction in HuR binding. Recently, Chk2 was shown to influence HuR function by regulating its phosphorylation in response to oxidative stress (Abdelmohsen et al., 2007). Consistent with results observed in HeLa cells, Chk2 also physically interacted with HuR in IEC-6 cells (Supplemental Figure A3). To investigate the possibility that polyamines modulated the association of HuR with c-Myc mRNA through Chk2-regulated HuR phosphorylation, we examined changes in Chk2 levels after decreased or increased cellular polyamines. As shown in Figure 5A, polyamine depletion by DFMO decreased the Chk2 abundance and inhibited its kinase activity as indicated by a decrease in levels of phosphorylated Chk2 (p-Chk2), whereas increasing cellular polyamines by ectopic ODC overexpression elevated the Chk2 protein levels and increased its kinase activity (Figure 5B). Interestingly, decreased levels of Chk2 after polyamine depletion were associated with a significant decrease in the level of phosphorylated HuR (p-HuR), although it failed to alter total HuR levels. Exogenous putrescine given together with DFMO prevented the reduction in Chk2 and also restored p-HuR to normal levels. Alternatively, induced Chk2 after increased polyamines by ODC overexpression resulted in an increase in the level of p-HuR. To further characterize the relationship between polyamine-regulated Chk2 and HuR phosphorylation, we determined the effect of Chk2 overexpression on the levels of p-HuR. As shown in Figure 5C, transient transfection with the Chk2 expression vector dramatically increased Chk2 protein expression levels (∼5-fold at 24 h and ∼12-fold at 48 h after transfection) compared with Chk2 levels in the population transfected with the control vector lacking Chk2 cDNA (Null). Increased Chk2 was associated with stimulation of HuR phosphorylation as indicated by an increase in p-HuR levels (Figure 5D). In addition, Chk2 overexpression did not induce the levels of whole-cell HuR.
Figure 5.

Polyamines regulate HuR phosphorylation through Chk2. (A) Representative immunoblots of total HuR (T-HuR), p-Chk2, and p-HuR after polyamine depletion. After cells were grown in control cultures and cultures containing DFMO alone or DFMO plus Put for 4 d, whole-cell lysates were harvested, and the levels of T-HuR, p-Chk2, and p-HuR were measured by Western blot analysis. To determine changes in levels of p-Chk2 and p-HuR, cell lysates were IPed by anti-Chk2 or anti-HuR antibody, and precipitates were analyzed by Western blotting with the antibody against all phosphorylated proteins (pProteins). (B) Changes in levels of T-HuR, p-Chk2, and p-HuR in control cells (vector alone) and two clones (C1 and C2) of stable cell lines overexpressing ODC (ODC-IEC). (C) Representative immunoblots of Chk2 protein. Cells were transfected with the Chk2 expression vector (Chk2) or control vector (Null) by using Lipofectamine. Whole-cell lysates were harvested at the indicated times after transfection, and Chk2 protein was measured by Western blot analysis. (D) Changes in levels of T-HuR and p-HuR proteins in cells transfected with the Chk2 or Null for 48 h. Three separate experiments were performed that showed similar results.
The results presented in Figure 6 further show that ectopic Chk2 overexpression not only increased the association of HuR with c-Myc mRNA in normal cells (without DFMO treatment) but also prevented the reduction in HuR binding in polyamine-deficient cells. The levels of [HuR/c-Myc mRNA] complexes in normal cells increased significantly after Chk2 overexpression as measured by biotin RNA pull-down (Figure 6Aa) and RNP IP (Figure 6B) assays. The induced HuR association with c-Myc mRNA by Chk2 overexpression was mediated by the c-Myc 3′-UTR, because transcripts corresponding to the coding region of c-Myc did not bind to HuR regardless of Chk2 expression levels (Figure 6, Ab and B, right). In polyamine-deficient cells, Chk2 overexpression also totally prevented the reduction in HuR binding to c-Myc mRNA (Figure 6, C and D). In fact, the levels of [HuR/c-Myc mRNA] complexes in polyamine-deficient cells after Chk2 overexpression were much higher than those observed in control cells (Figure 6, C and D). In addition, ectopic Chk2 overexpression did not alter the levels of total c-Myc RNA in polyamine-deficient cells (data not shown). Together, these findings suggest that polyamines regulate the interaction of HuR with the c-Myc 3′-UTR through Chk2-mediated HuR phosphorylation.
Figure 6.
Ectopic Chk2 overexpression increases HuR binding to c-Myc mRNA. (A) Representative immunoblots of HuR by using the pull-down materials from the c-Myc 3′-UTR (a) or CR (b) in control cells. Cytoplasmic lysates prepared from cells transfected with the Chk2 or Null for 48 h were incubated with biotinylated c-Myc 3′-UTR or CR for 30 min at 25°C, and the resulting RNP complexes were pulled down by using streptavidin-coated beads. Levels of HuR protein in the pull-down materials were measured by Western blot analysis. (B) Association of endogenous HuR with endogenous c-Myc mRNA in cells described in A. Whole lysates from controls and Chk2-transfected cells were used for IP in the presence of anti-HuR antibody or nonspecific IgG. RNA in the IP material was used in RT-qPCR reactions to detect the presence of c-Myc mRNA and the resulting PCR products (∼249 base pairs) were visualized in agarose gels. (C) Representative immunoblots of HuR protein using the pull-down materials from the c-Myc 3′-UTR (a) or CR (b) in polyamine-deficient cells. After cells were treated with DFMO for 2 d, they were transfected with the Chk2 or Null and cultured for additional 48 h in the presence of DFMO. HuR binding to c-Myc mRNA was examined by biotin pull-down assays. (D) Association of endogenous HuR with endogenous c-Myc mRNA in cells described in C. Three separate experiments were performed that showed similar results.
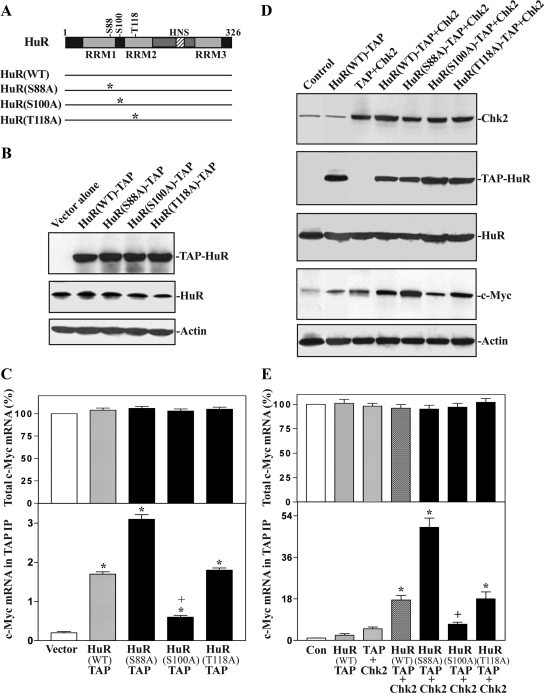
Point Mutation at Chk2 Phosphorylation Sites Alters HuR Binding to c-Myc mRNA
Chk2 has shown to phosphorylate HuR at residues S88, S100, and T118, but each individual phosphorylation site plays a distinct role in regulating HuR binding to different target mRNAs (Abdelmohsen et al., 2007). In this study, HuR mutants with alanine substitutions at each of the predicted Chk2 phosphorylation sites were tested. As shown in Figure 7, B and C, ectopic expression of wild-type (WT) HuR-TAP fusion protein increased the association of HuR with c-Myc mRNA, whereas point mutation at Chk2 phosphorylation sites modulated HuR binding to c-Myc mRNA. Importantly, point mutations at each individual Chk2 phosphorylation site in HuR had a distinct effect on the binding of the HuR-TAP proteins to c-Myc mRNA (Figure 7C, bottom), although they did not affect the levels of total c-Myc mRNA (Figure 7C, top). Cells transfected with the HuR(S88A)-TAP exhibited an increase in levels of c-Myc mRNA associated with the chimeric protein when compared with those obtained from cells transfected with the HuR(WT)-TAP, whereas cells overexpressing HuR(S100A)-TAP displayed a decreased association with the c-Myc mRNA. T118A mutation did not affect its binding to c-Myc mRNA, as the levels of the complex were similar in the HuR(T118A)-TAP and HuR(WT)-TAP groups. We also examined the effect of cotransfection of the Chk2 expression vector with different HuR-TAP mutants on HuR association with c-Myc mRNA and demonstrated that point mutation at Chk2 phosphorylation sites in HuR also altered Chk2-induced HuR binding to c-Myc mRNA, thus affecting c-Myc translation. The results presented in Figure 7D show that Chk2 overexpression alone slightly increased c-Myc protein expression level (∼2-fold) compared with those observed in control cells, but cotransfection of Chk2 with HuR(WT)-TAP remarkably increased levels of c-Myc protein (∼6-fold). Neither Chk2 overexpression nor HuR overexpression altered the stability of c-Myc protein (Supplemental Fig. A4). When the association of HuR with the c-Myc mRNA was studied, the level of the complexes increased marginally in cells transfected with Chk2 alone but was remarkably elevated in cells cotransfected with Chk2 and HuR(WT)-TAP (Figure 7E, bottom). In contrast, cells in the Chk2 and HuR(S88A)-TAP transfection group exhibited an increase in the association of c-Myc mRNA with the chimeric protein when compared with cells in the Chk2 and HuR(WT)-TAP group, whereas cells overexpressing Chk2 and HuR(S100A)-TAP showed decreased association with the c-Myc mRNA. T118A mutation did not affect Chk2-indiced HuR association with c-Myc mRNA. In addition, cotransfection of the Chk2 expression vector with either HuR(WT)-TAP or different HuR-TAP mutants failed to alter total c-Myc mRNA levels (Figure 7E, top). Consistently, c-Myc protein expression increased in cells expressing Chk2+HuR(S88A)-TAP, decreased in cells expressing Chk2+HuR(S100A)-TAP, and showed no change in cells expressing Chk2+HuR(T118A)-TAP, compared with those observed in cells overexpressing Chk2 and HuR(WT)-TAP (Figure 7D, bottom). Together, these findings indicate that phosphorylation at residue S100 by Chk2 enhances the affinity of HuR for c-Myc mRNA and is implicated in regulating c-Myc translation.
Figure 7.
Ectopic overexpression of HuR-carrying point mutations alters c-Myc expression by altering [HuR/c-Myc mRNA] association. (A) Schematic of point mutations introduced at the three predicted residues of phosphorylation by Chk2. (B) Changes in levels of HuR-TAP fusion proteins (TAP-HuR) and endogenous HuR. Cells were transfected with the vector expressing wild-type (WT) HuR-TAP or mutated HuR-TAP fusion proteins. The levels of chimeric TAP-HuR and endogenous HuR were measured by Western immunoblotting analysis 48 h after the transfection using specific antibody against TAP or HuR. Three separate experiments were performed that showed similar results. (C) Changes in levels of the total c-Myc mRNA (top) and c-Myc mRNA in TAP IP materials (bottom) in cells described in B. Total c-Myc mRNA levels were measured by RT-qPCR analysis, whereas binding of chimeric HuR-TAP proteins to c-Myc mRNA was examined by performing TAP IP followed by real-time quantitative PCR analysis. Values are means ± SE of data from three separate experiments. *p and +p < 0.05 compared with controls (vector) and cells transfected with the HuR(WT)-TAP, respectively. (D) Changes in levels of c-Myc protein after overexpression of Chk2 and chimeric TAP-HuR proteins. Cells were cotransfected with the Chk2 expression vector (Chk2) and the vector expressing HuR(WT)-TAP or mutated HuR-TAP fusion proteins. The levels of Chk2, TAP-HuR, endogenous HuR, and c-Myc proteins were measured 48 h after the transfection. (E) Changes in levels of the total c-Myc mRNA (top) and c-Myc mRNA in TAP IP materials (bottom) in cells described in D as measured by real-time quantitative PCR analysis. *p < 0.05 compared with controls (Con) and cells transfected with Chk2; +p < 0.05 compared with cells cotransfected with Chk2 and WT-HuR.
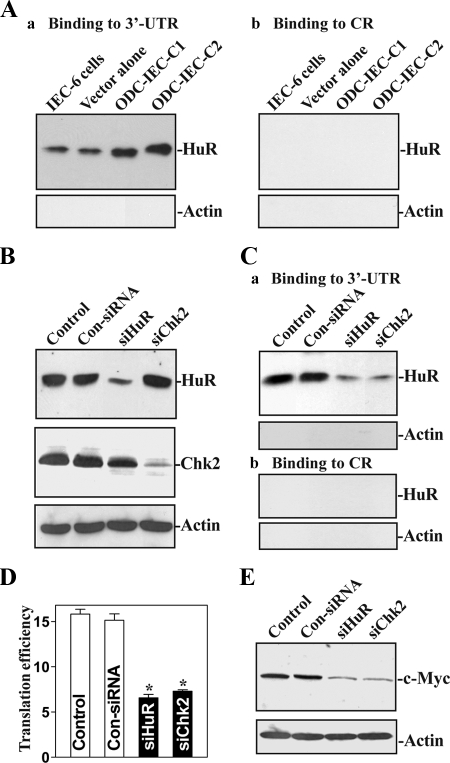
Silencing HuR or Chk2 Abolishes the Increase in c-Myc Translation by Polyamines
The results presented in Figure 8A show that [HuR/c-Myc mRNA] complexes increased in cells stably overexpressing ODC relative to parental IEC-6 cells and to cells transfected with the empty vector alone. To reduce HuR and Chk2 levels, small interfering RNAs (siRNAs) targeting the coding regions of HuR mRNA (siHuR) or Chk2 mRNA (siChk2) were used, and the putative roles of HuR and Chk2 in the stimulation of c-Myc mRNA translation after increasing cellular polyamines were examined. With >95% cells transfected (data not shown), transfection with siHuR or Chk2 potently and specifically silenced HuR or Chk2 expression in stable ODC-IEC cells (Figure 8B). Silencing HuR or Chk2 not only decreased [HuR/c-Myc mRNA] association (Figure 8C) but also inhibited c-Myc mRNA translation, as indicated by a decrease in levels of c-Myc ARE luciferase reporter gene activity (Figure 8D). Analysis of the changes in half-life of luciferase reporter mRNA after HuR silencing revealed no significant changes in luciferase reporter mRNA stability in HuR-silenced populations (data not shown). Consistently, in HuR- or Chk2-silenced populations, the c-Myc protein levels were also decreased (Figure 8E). In contrast, transfection with control (Con)-siRNA had no effects on HuR binding to c-Myc 3′-UTR, c-Myc mRNA translation, or c-Myc protein levels. These findings strongly suggest that Chk2-induced HuR phosphorylation is necessary for the stimulation of c-Myc mRNA translation after induction in cellular polyamines, in turn elevating c-Myc expression and thereby inducing cell proliferation.
Figure 8.
Effects of silencing HuR and Chk2 on [HuR/c-Myc mRNA] association and c-Myc translation in stable ODC-expressing cells (ODC-IEC). (A) Changes in HuR binding to c-Myc mRNA in parental IEC-6 cells, IEC-6 cells transfected with the empty vector alone, and two clones (C) of stable transfected ODC-IEC cells as detected by biotin pull-down assays: a, binding to 3′-UTR and b, binding to CR. Three independent experiments were performed showing similar results. (B) Representative HuR and Chk2 immunoblots. Stable ODC-IEC cells were transfected with either siRNA targeting the HuR mRNA coding region (siHuR), siRNA targeting the Chk2 mRNA coding region (siChk2), or control-siRNA (Con-siRNA), and whole-cell lysates were harvested 48 h thereafter. The levels of HuR and Chk2 proteins were measured by Western blot analysis, and equal loading was monitored by assessing β-actin levels. (C) HuR binding to c-Myc mRNA as detected by biotin pull down assays in cells described in B: a, binding to 3′-UTR and b, binding to CR. (D) Changes in c-Myc translation efficiency as measured by using pGL3-Luc-MycARE reporter assays in cells described in B. Twenty-four hours after transfection of cells with siHuR or siChk2 for 24 h, they were transfected with the pGl3-Luc-MycARE or pGL3-Luc (negative control). Transfected cells were harvested and assayed for luciferase activity 24 h after transfection with the pGl3-Luc-MycARE or pGL3-Luc. Data were normalized by Renilla-driven luciferase activity and expressed as means ± SE of data from three separate experiments. *p < 0.05 compared with control and cells transfected with Con-siRNA. (E) Changes in expression of c-Myc protein as measured by Western blot analysis of cells described in B. Data are representative from three independent experiments showing similar results.
DISCUSSION
The transcriptional regulation of c-Myc expression by polyamines is well documented (Celano et al., 1989; Patel and Wang, 1997), but little is known about the role and mechanism of cellular polyamines in the control of c-Myc translation. In the present study, we identified a novel function of polyamines in the regulation of c-Myc mRNA translation, thus advancing our understanding of the biological functions of cellular polyamines. Experiments aimed at characterizing the molecular aspects of this process suggested that polyamines regulated HuR association with the c-Myc mRNA through Chk2-regulated HuR phosphorylation, probably at HuR(S100). Increasing cellular polyamines induced Chk2 activity, increased the abundance of phosphorylated HuR, and enhanced the affinity of HuR for c-Myc mRNA, thereby stimulating c-Myc translation. Conversely, polyamine depletion reduced Chk2-dependent HuR phosphorylation and lowered [HuR-c-Myc mRNA] associations, leading to a reduction in c-Myc protein synthesis.
c-Myc Translational Regulation by Polyamines
The human c-Myc transcripts are generated from four alternative promoters (P0, P1, P2, and P3), but most transcription of c-Myc mRNA in normal cells is directed by the P2 promoter (ar-Rushdi et al., 1983). P2 mRNA governs translation of two distinct Myc proteins of 67 and 64 kDa (designated c-Myc1 and c-Myc2) by an alternative initiation mechanism involving two in-frame start codons, CUG and AUG, respectively (Hann et al., 1988). Although c-Myc expression is crucially modulated by transcription, the strong contribution of its posttranscriptional regulation, especially c-Myc mRNA translation, is increasingly recognized (Kim et al., 2003; Liao et al., 2007; Wall et al., 2008). Several studies have demonstrated that the specific RNA sequences located in the c-Myc 5′ and 3′-UTRs are intimately implicated in the c-Myc translational control (Stoneley et al., 2000; Liao et al., 2007; Wall et al., 2008). For example, the 5′-UTR of c-Myc contains an internal ribosomal entry site (IRES) that lies in the 5′-UTR upstream of the AUG start codon of the major c-Myc protein (c-Myc2) (Nanbru et al., 1997; Stoneley et al., 1998). Moreover, translation of c-Myc mRNA occurs via a cap-dependent mechanism as well as an IRES-dependent mechanism (Stoneley et al., 2000). In contrast, the c-Myc 3′-UTR also contains abundant AREs that directly interact with RBPs such as HuR, AUF1, and TIA-1–related protein (TIAR) (Lafon et al., 1998). The interactions of RPBs with the c-Myc AREs play a critical role in the control of c-Myc mRNA translation. Recently, it was reported that AUF1 and TIAR competitively bind to the AREs of c-Myc 3′-UTR and that the ratios of AUF1 and TIAR bound to c-Myc mRNA permits the dynamic control of c-Myc translation and cell proliferation (Liao et al., 2007).
In virtually all published studies, depletion of cellular polyamines represses c-Myc expression (Celano et al., 1989; Patel and Wang, 1997; Gerner and Meyskens, 2004; Casero and Marton, 2007), with only one exception, F9 teratocarcinoma stem cells, where polyamine depletion up-regulates c-Myc expression (Frostesjo and Heby, 1999). Although the exact reasons why c-Myc levels increase in F9 cells after polyamine depletion remain unknown, this response is cell-type dependent and may be related to the distinct function of c-Myc and the mechanisms that control c-Myc expression in this particular line of cancer cells. In this regard, polyamine depletion induces both terminal differentiation and G1 growth arrest in F9 cells (Frostesjo and Heby, 1999), but it only results in growth arrest without affecting differentiation in IECs (Wang et al., 1993; Li et al., 1999). Consistently, ectopic c-Myc overexpression enhances IEC-6 cell proliferation but has no effect on cell differentiation (Liu et al., 2005). The results reported in the present study show that alterations of the intracellular polyamine levels potently change the translational efficiency of c-Myc mRNA in IECs and that this regulatory effect is mediated through the c-Myc 3′-UTR. Polyamine depletion repressed c-Myc mRNA translation as indicated by a decrease in the levels of newly synthesized c-Myc protein and c-Myc-ARE luciferase reporter gene activity (Figure 1), whereas elevated polyamine levels enhanced c-Myc translation via the c-Myc ARE (Figure 2). Data from polysome analysis further show that polyamine depletion caused a shift in the distribution of c-Myc mRNA toward nontranslating or low-translating fractions of the gradient, and that this shift was completely prevented by exogenous addition of the polyamine putrescine. Consistent with our observations, polyamines were also shown to stimulate translation of the spermidine-spermine N1-acetyltransferase (SSAT) mRNA by inhibiting the binding of repressor RBPs to the SSAT transcript, but the specific RBPs involved in this process remain to be elucidated (Butcher et al., 2007). In addition, polyamines are shown to increase the unproductive splicing of SSAT mRNA; this action also contributes to the induction in SSAT translation by polyamines (Hyvönen et al., 2006). We recently demonstrated that polyamines modulate the translation of a tight junction protein, zona occludens-1 (ZO-1), and that depletion of cellular polyamines represses ZO-1 translation by increasing TIAR association with the ZO-1 3′-UTR (Chen et al., 2008).
Polyamines Are Necessary for HuR Association with c-Myc mRNA
Our results also indicate that polyamines are necessary for HuR binding to the c-Myc 3′-UTR that contains AU- and U-rich stretches (Figure 4A). Polyamine depletion resulted in HuR dissociation from the c-Myc mRNA, although cytoplasmic HuR levels were increased in this condition. Because HuR association with specific target transcripts is tightly regulated in response to cellular stress and that HuR functions as a translational enhancer in several instances (reviewed in Abdelmohsen et al., 2007), these observations support the hypothesis that increasing polyamines can stimulate c-Myc translation by inducing [HuR-c-Myc mRNA] associations, whereas depleting polyamines can inhibit c-Myc translation by repressing the interaction of HuR with c-Myc mRNA. The results shown in Figure 8 further support this notion, as HuR silencing or silencing Chk2 inhibited c-Myc translation and reduced c-Myc protein levels in cells overexpressing ODC. An increasing body of evidence indicates that the changes in HuR association to a given mRNA are dependent on the transcript itself rather than the particular stimulus (Zou et al., 2006; Abdelmohsen et al., 2007; Xiao et al., 2007). Our previous studies showed that polyamine depletion enhanced HuR association with mRNAs encoding p53, NPM, and ATF2 and stabilized these transcripts (Zou et al., 2006; Wang, 2007; Xiao et al., 2007). In another study, endoplasmic reticulum stress caused a transient dissociation of HuR from the cytochrome c mRNA, whereas it promoted the binding of a translational repressor, TIA-1 (Kawai et al., 2006). Recently, other factors such as microRNAs were also shown to influence the association of HuR with target mRNAs. For example, it was reported that the association of microRNA miR-122 with the CAT-1 mRNA represses its translation (Bhattacharyya et al., 2006). In cells exposed to stress agents, HuR associated with the CAT-1 3′-UTR, thereby interfering with the binding of miR-122, and relieving the miR-122-imposed translation repression. Kim et al. (2009) recently demonstrated that HuR recruits let-7/RISC complex to repress c-Myc expression in HeLa cells, suggesting that the regulatory effect of HuR on c-Myc translation could be influenced by a the microRNA let-7. By contrast, IEC-6 cells did not express detectable levels of let-7, regardless of the presence or absence of cellular polyamines (data not shown). The effects of cellular polyamines on the expression and function of microRNAs and RBPs that repress translation are the focus of ongoing studies.
Polyamines Regulate HuR Phosphorylation through Chk2
It is interesting to note that although the cytoplasmic HuR levels acutely increase in polyamine-deficient cells (Figure 4; Zou et al., 2006), HuR association with the c-Myc mRNA is significantly decreased, suggesting that posttranslational events are implicated in triggering this reduction in HuR binding. HuR was recently reported to be phosphorylated by Chk2 kinase in vivo as well as in vitro and this posttranslational modification influenced HuR's association with target transcripts (Abdelmohsen et al., 2007). Many substrates and interacting partners of Chk2 have since been identified and through this increasingly diverse array of interactions, Chk2 is shown to act not only as a regulator of DNA damage-response signaling and activator of cell cycle checkpoint, but also of apoptosis, senescence, viral infectivity, and other pathways (Pommier et al., 2006; Li et al., 2008). Although cellular polyamines seem to affect Chk2 levels and activity, they do not seem to be directly involved in the DNA damage response (Casero and Marton, 2007; Gerner and Meyskens, 2004). In this study, we demonstrated that Chk2 expression requires polyamines and that polyamines regulate HuR phosphorylation by altering Chk2 levels. Depletion of cellular polyamines reduced Chk2 activity and decreased p-HuR levels, associated with a reduction in [HuR-c-Myc mRNA] complexes. Ectopic Chk2 overexpression increased HuR phosphorylation, thus enhancing HuR association with the c-Myc mRNA in normal and polyamine-deficient cells. In fact, the levels of [HuR-c-Myc mRNA] complexes in polyamine-deficient cells are markedly higher than those observed in control cells after Chk2 overexpression (Figure 6). This induction in HuR binding to c-Myc mRNA was not surprising, as polyamine-deficient cells showed higher cytoplasmic HuR levels. These findings suggest a novel function whereby polyamines regulate c-Myc translation by modulating Chk2 levels.
Although the exact role of HuR phosphorylation at the putative Chk2 target residues remains to be fully elucidated, studies using various HuR mutations suggested that HuR phosphorylation at residues S88 and S100 by Chk2 critically modulated [HuR-c-Myc mRNA] association (Figure 7). However, HuR mutants lacking a given Chk2 phosphorylation site displayed distinct binding affinities for c-Myc mRNA: S88A mutant bound c-Myc mRNA more effectively than WT, whereas S100A mutant associated with the c-Myc transcript less than WT did. Consistent with this pattern of interactions, cells overexpressing the S88A mutant exhibited an increase in c-Myc protein expression levels, whereas overexpression of the S100A mutant decreased c-Myc abundance (Figure 7D). In contrast, T118A mutant did not show altered interaction with the c-Myc mRNA nor did it cause differences in c-Myc protein levels. S88 and T118 lie within RRM1 and RRM2, respectively, and S100 lies between RRM1 and RRM2. It is unclear at present why HuR phosphorylation at different sites by Chk2 displays variable effects on its affinity for target transcripts, but it could alter the relative position of RRM1 and RRM2. Given these distinct effects, double and triple nonphosphorylatable HuR mutants were not studied, as their effects would be difficult to interpret. Further studies, including mass spectroscopy and crystallographic analysis, are needed for solving this important question.
Finally, the data obtained in the present study suggest that polyamine-regulated c-Myc translation is physiologically significant because polyamines are essential for maintaining intestinal epithelial integrity and because cellular polyamine levels are highly regulated by epithelial growth status and by stress stimuli (Seiler and Raul, 2007). Increasing the levels of cellular polyamines during normal gut mucosal growth and in regenerating mucosa after injury stimulates epithelial cell renewal by inducing c-Myc expression (Wang et al., 1993; Wang and Johnson, 1994; Liu et al., 2006); polyamine depletion causes mucosal atrophy and delays mucosal healing by reducing c-Myc expression levels (Wang et al., 1991). The current work indicates that polyamines enhance HuR association with the c-Myc mRNA through Chk2-mediated HuR phosphorylation, thus implicating c-Myc in this important biological process. Furthermore, HuR silencing or inhibition of HuR phosphorylation by Chk2 reduction repressed c-Myc translation and decreased c-Myc protein levels in cells overexpressing ODC. These findings provide novel evidence that polyamines are required for the stimulation of normal cell growth and proliferation by inducing c-Myc translation through HuR.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Merit Review grant (to J.-Y.W.) from the Department of Veterans Affairs and by National Institutes of Health grants DK-57819, DK-61972, and DK-68491 (to J.-Y.W.). J.-Y.W. is a Research Career Scientist, Medical Research Service, U.S. Department of Veterans Affairs. M. G. is supported by the National Institute on Aging-Intramural Research Program, National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-07-0550) on October 7, 2009.
REFERENCES
- Abdelmohsen K., et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelmohsen K., Kuwano Y., Kim H. H., Gorospe M. Posttranscriptional gene regulation by RNA-binding proteins during oxidative stress: implications for cellular senescence. Biol. Chem. 2008;389:243–255. doi: 10.1515/BC.2008.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ar-Rushdi A., Nishikura K., Erikson J., Watt R., Rovera G., Croce C. M. Differential expression of the translocated and the untranslocated c-myc oncogene in Burkitt lymphoma. Science. 1983;222:390–393. doi: 10.1126/science.6414084. [DOI] [PubMed] [Google Scholar]
- Bakheet T., Frevel M., Williams B. R., Greer W., Khabar K. S. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belting M., Borsig L., Fuster M. M., Brown J. R., Persson L., Fransson L. A., Esko J. D. Tumor attenuation by combined heparan sulfate and polyamine depletion. Proc. Natl. Acad. Sci. USA. 2002;99:371–376. doi: 10.1073/pnas.012346499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S. N., Habermacher R., Martine U., Closs E. I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Butcher N. J., Broadhurst G. M., Minchin R. F. Polyamine-dependent regulation of spermidine-spermine N1-acetyltransferase mRNA translation. J. Biol. Chem. 2007;282:28530–28539. doi: 10.1074/jbc.M701265200. [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai W. S., Blackshear P. J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- Casero R. A., Jr., Marton L. J. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007;6:373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- Celano P., Baylin S. B., Casero R. A., Jr. Polyamines differentially modulate the transcription of growth-associated genes in human colon carcinoma cells. J. Biol. Chem. 1989;264:8922–8927. [PubMed] [Google Scholar]
- Chen C. Y., Shyu A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chen J., Xiao L., Rao J. N., Zou T., Liu L., Bellavance E., Gorospe M., Wang J. Y. JunD represses transcription and translation of the tight junction protein zona occludens-1 modulating intestinal epithelial barrier function. Mol. Biol. Cell. 2008;19:3701–3712. doi: 10.1091/mbc.E08-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A., Huwiler A., Müller R., Radeke H. H., Pfeilschifter J., Eberhardt W. Protein kinase Cα-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol. Biol. Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin. Cell Dev. Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fan X. C., Steitz J. A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frostesjo L., Heby O. Polyamine depletion up-regulates c-Myc expression, yet induces G1 arrest and terminal differentiation of F9 teratocarcinoma stem cells. J. Cell Biochem. 1999;76:143–152. doi: 10.1002/(sici)1097-4644(20000101)76:1<143::aid-jcb14>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Gerner E. W., Meyskens F. L. Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer. 2004;4:781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- Gherzi R., Lee K. Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C. Y. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Ghoda L., van Daalen Wetters T., Macrae M., Ascherman D., Coffino P. Prevention of rapid intracellular degradation of ODC by a carboxyl-terminal truncation. Science. 1989;243:1493–1495. doi: 10.1126/science.2928784. [DOI] [PubMed] [Google Scholar]
- Gorospe M. HuR in the mammalian genotoxic response: post-transcriptional multitasking. Cell Cycle. 2003;2:412–414. [PubMed] [Google Scholar]
- Hann S. R., King M. W., Bentley D. L., Anderson C. W., Eisenman R. N. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt's lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Harter J. L. Critical values for Duncan's new multiple range test. Biometrics. 1960;16:671–685. [Google Scholar]
- Hinman M. N., Lou H. Diverse molecular functions of Hu proteins. Cell Mol. Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyvönen M. T., et al. Polyamine-regulated unproductive splicing and translation of spermidine/spermine N1-acetyltransferase. RNA. 2006;12:1569–1582. doi: 10.1261/rna.39806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Lal A., Yang X., Galban S., Mazan-Mamczarz K., Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol. Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fitzler M. J., Scheuner D., Kaufman R. J., Golan D. E., Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Kim H. H. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23:1743–1748. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Paek K. Y., Choi K., Kim T. D., Hahm B., Kim K. T., Jang S. K. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell Biol. 2003;23:708–720. doi: 10.1128/MCB.23.2.708-720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer D. L., Chang B. D., Chen Y., Diegelman P., Alm K., Black A. R., Roninson I. B., Porter C. W. Polyamine depletion in human melanoma cells leads to G1 arrest associated with induction of p21WAF1/CIP1/SDI1, changes in the expression of p21-regulated genes, and a senescence-like phenotype. Cancer Res. 2001;61:7754–7762. [PubMed] [Google Scholar]
- Lafon I., Carballès F., Brewer G., Poiret M., Morello D. Developmental expression of AUF1 and HuR, two c-myc mRNA binding proteins. Oncogene. 1998;16:3413–3421. doi: 10.1038/sj.onc.1201895. [DOI] [PubMed] [Google Scholar]
- Laroia G., Cuesta R., Brewer G., Schneider R. J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- Li L., Li J., Rao J. N., Li M., Bass B. L., Wang J. Y. Inhibition of polyamine synthesis induces p53 gene expression but not apoptosis. Am. J. Physiol. Cell Physiol. 1999;276:C946–C954. doi: 10.1152/ajpcell.1999.276.4.C946. [DOI] [PubMed] [Google Scholar]
- Li L., Liu L., Rao J. N., Esmaili A., Strauch E. D., Bass B. L., Wang J. Y. JunD stabilization results in inhibition of normal intestinal epithelial cell growth through p21 after polyamine depletion. Gastroenterology. 2002;123:764–779. doi: 10.1053/gast.2002.35386. [DOI] [PubMed] [Google Scholar]
- Li L., Rao J. N., Guo X., Liu L., Santora R., Bass B. L., Wang J. Y. Polyamine depletion stabilizes p53 resulting in inhibition of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 2001;281:C941–C953. doi: 10.1152/ajpcell.2001.281.3.C941. [DOI] [PubMed] [Google Scholar]
- Li J., Taylor I. A., Lloyd J., Clapperton J. A., Howell S., MacMillan D., Smerdon S. J. Chk2 oligomerization studied by phosphopeptide ligation: implication for regulation and phosphodependent interactions. J. Biol. Chem. 2008;283:36019–36030. doi: 10.1074/jbc.M804075200. [DOI] [PubMed] [Google Scholar]
- Liao B., Hu Y., Brewer G. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 2007;14:511–518. doi: 10.1038/nsmb1249. [DOI] [PubMed] [Google Scholar]
- Liu L., Guo X., Rao J. N., Zou T., Marasa B. S., Chen J., Greenspon J., Casero R. A., Jr., Wang J. Y. Polyamine-modulated c-myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem. J. 2006;398:257–267. doi: 10.1042/BJ20060217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Li L., Rao J. N., Zou T., Zhang H. M., Boneva D., Bernard M. S., Wang J. Y. Polyamine-modulated expression of c-myc plays a critical role in stimulation of normal intestinal epithelial cell proliferation. Am. J. Physiol. Cell Physiol. 2005;288:C89–C99. doi: 10.1152/ajpcell.00326.2004. [DOI] [PubMed] [Google Scholar]
- López de Silanes I., Lal A., Gorospe M. HuR: post-transcriptional paths to malignancy. RNA Biol. 2004a;1:135–137. doi: 10.4161/rna.2.1.1552. [DOI] [PubMed] [Google Scholar]
- López de Silanes I., Zhan M., Lal A., Yang X., Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA. 2004b;101:2987–2992. doi: 10.1073/pnas.0306453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcu K. B., Bossone S. A., Patel A. J. myc function and regulation. Annu. Rev. Biochem. 1992;61:809–860. doi: 10.1146/annurev.bi.61.070192.004113. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Galban S., de Silanes I. L., Martindale J. L., Atasoy U., Keene J. D., Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc. Natl. Acad. Sci. USA. 2003;100:8354–8359. doi: 10.1073/pnas.1432104100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack S. A., Johnson L. R. Role of polyamines in gastrointestinal mucosal growth. Am. J. Physiol. Gastrointest. Liver Physiol. 1991;260:G795–G806. doi: 10.1152/ajpgi.1991.260.6.G795. [DOI] [PubMed] [Google Scholar]
- Nanbru C., Lafon I., Audigier S., Gensac M. C., Vagner S., Huez G., Prats A. C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 1997;272:32061–32063. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- Paasinen-Sohns A., Kielosto M., Kääriäinen E., Eloranta T., Laine A., Jänne O. A., Birrer M. J., Hölttä E. c-Jun activation-dependent tumorigenic transformation induced paradoxically by overexpression or block of S-adenosylmethionine decarboxylase. J. Cell Biol. 2000;151:801–809. doi: 10.1083/jcb.151.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. R., Wang J. Y. Polyamines modulate transcription but not posttranscription of c-myc and c-jun in IEC-6 cells. Am. J. Physiol. Cell Physiol. 1997;273:C1020–C1029. doi: 10.1152/ajpcell.1997.273.3.C1020. [DOI] [PubMed] [Google Scholar]
- Patel A. R., Wang J. Y. Polyamine depletion is associated with an increase in JunD/AP-1 activity in small intestinal crypt cells. Am. J. Physiol. Gastrointest. Liver Physiol. 1999;276:G441–G450. doi: 10.1152/ajpgi.1999.276.2.G441. [DOI] [PubMed] [Google Scholar]
- Patel A. R., Li J., Bass B. L., Wang J. Y. Expression of the transforming growth factor-beta gene during growth inhibition following polyamine depletion. Am. J. Physiol. Cell Physiol. 1998;275:C590–C598. doi: 10.1152/ajpcell.1998.275.2.C590. [DOI] [PubMed] [Google Scholar]
- Pommier Y., Weinstein J. N., Aladjem M. I., Kohn K. W. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Mol. Pathw. 2006;12:2657–2661. doi: 10.1158/1078-0432.CCR-06-0743. [DOI] [PubMed] [Google Scholar]
- Quaroni A., Wands J., Trelstad R. L., Isselbacher K. J. Epithelioid cell cultures from rat small intestine: characterization by morphologic and immunologic criteria. J. Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray R. M., Zimmerman B. J., McCormack S. A., Patel T. B., Johnson L. R. Polyamine depletion arrests cell cycle and induces inhibitors p21(Waf1/Cip1), p27(Kip1), and p53 in IEC-6 cells. Am. J. Physiol. Cell Physiol. 1999;276:C684–C691. doi: 10.1152/ajpcell.1999.276.3.C684. [DOI] [PubMed] [Google Scholar]
- Ross J. mRNA stability in mammalian cells. Microbiol. Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. M., Birnie G. D. Myc oncogenes: the enigmatic family. Biochem. J. 1996;314:713–721. doi: 10.1042/bj3140713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler N., Raul F. Polyamines and the intestinal tract. Crit. Rev. Clin. Lab. Sci. 2007;44:365–411. doi: 10.1080/10408360701250016. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Paulin F. E., Le Quesne J. P., Chappell S. A., Willis A. E. c-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Stoneley M., Subkhankulova T., Le Quesne J. P., Coldwell M. J., Jopling C. L., Belsham G. J., Willis A. E. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., Bishop J. M. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Wall M., Poortinga G., Hannan K. M., Pearson R. B., Hannan R. D., McArthur G. A. Translational control of c-MYC by rapamycin promotes terminal myeloid differentiation. Blood. 2008;112:2305–2317. doi: 10.1182/blood-2007-09-111856. [DOI] [PubMed] [Google Scholar]
- Wang J. Y. Polyamines and mRNA stability in regulation of intestinal mucosal growth. Amino Acids. 2007;33:241–252. doi: 10.1007/s00726-007-0518-z. [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Johnson L. R. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology. 1991;100:333–343. doi: 10.1016/0016-5085(91)90200-5. [DOI] [PubMed] [Google Scholar]
- Wang J. Y., Johnson L. R. Expression of protooncogenes c-fos and c-myc in healing of gastric mucosal stress ulcers. Am. J. Physiol. Gastrointest. Liver Physiol. 1994;266:G878–G886. doi: 10.1152/ajpgi.1994.266.5.G878. [DOI] [PubMed] [Google Scholar]
- Wang J. Y., McCormack S. A., Viar M. J., Johnson L. R. Stimulation of proximal small intestinal mucosal growth by luminal polyamines. Am. J. Physiol. Gastrointest. Liver Physiol. 1991;261:504–511. doi: 10.1152/ajpgi.1991.261.3.G504. [DOI] [PubMed] [Google Scholar]
- Wang J. Y., McCormack S. A., Viar M. J., Wang H., Tzen C. Y., Scott R. E., Johnson L. R. Decreased expression of protooncogenes c-fos, c-myc, and c-jun following polyamine depletion in IEC-6 cells. Am. J. Physiol. Gastrointest. Liver Physiol. 1993;265:G331–G338. doi: 10.1152/ajpgi.1993.265.2.G331. [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Wiluszm J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Wormington M., Peltz S. W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Xiao L., Rao J. N., Zou T., Liu L., Marasa B. S., Chen J., Turner D. J., Zhou H., Gorospe M., Wang J. Y. Polyamines regulate the stability of activating transcription factor-2 mRNA through RNA-binding protein HuR in intestinal epithelial cells. Mol. Biol. Cell. 2007;18:4579–4590. doi: 10.1091/mbc.E07-07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A. H., Rao J. N., Zou T., Liu L., Marasa B. S., Xiao L., Chen J., Turner D. J., Wang J. Y. p53-dependent NDRG1 expression induces inhibition of intestinal epithelial cell proliferation but not apoptosis after polyamine depletion. Am. J. Physiol. Cell Physiol. 2007;293:C379–C389. doi: 10.1152/ajpcell.00547.2006. [DOI] [PubMed] [Google Scholar]
- Zou T., Liu L., Rao J. N., Marasa B. S., Chen J., Xiao L., Zhou H., Gorospe M., Wang J. Y. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin α1. Biochem. J. 2008;409:389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T., Mazan-Mamczarz K., Rao J. N., Liu L., Marasa B. S., Zhang A. H., Xiao L., Pullmann R., Gorospe M., Wang J. Y. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J. Biol. Chem. 2006;281:19387–19394. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- Zou T., Rao J. N., Liu L., Marasa B. S., Keledjian K. M., Zhang A. H., Xiao L., Bass B. L., Wang J. Y. Polyamine depletion induces nucleophosmin modulating stability and transcriptional activity of p53 in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2005;289:C686–C696. doi: 10.1152/ajpcell.00085.2005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.