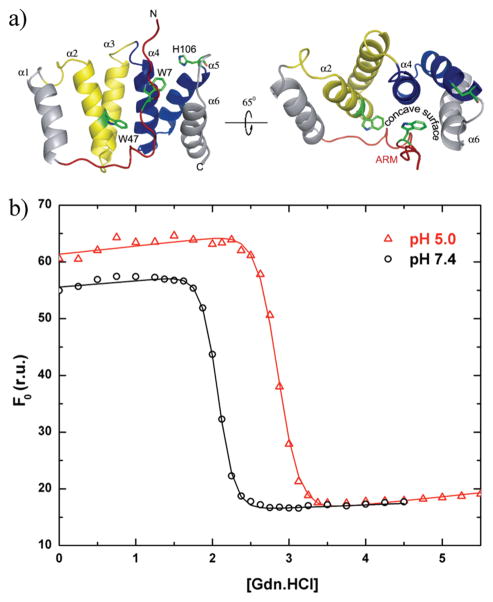
Figure 1.
Thermodynamic stability of Fis1ΔTM, which is lower at pH 7.4 than at pH 5.0 as measured by equilibrium chemical denaturation experiments. (a) Ribbon model depicting the NMR-derived structure of the cytosolic domain of Fis1 atpH 5.5 and 32°C, which displays a 16-residue N-terminal region (the Fis1 arm) draped over a concave surface created by α-helices 2, 4, and 6 [PDB entry 1y8m (29)]. The structure is color-coded in this and other figures to highlight the Fis1 arm (red) and the first (yellow) and second (blue) tetratricopeptide repeats. (b) Chemical denaturation experiments in which the unfolding reaction was followed as a function of increasing GdnHCl concentration by measuring the intrinsic tryptophan fluorescence of Trp7 and Trp47 at 325 nm upon excitation at 295 nm. The resulting data were fit to a two-state model using the method of Santoro and Bolen (56). The free energy of unfolding (ΔG)equals 12.1 ± 0.9 kcal/mol at pH 5.0 and 10.4 ± 1.1 kcal/mol at pH 7.4 (1.7 kcal/mol lower). Note the differences and similarities in the baselines for the folded and unfolded states, respectively.