Abstract
Purpose
The metastatic tumor antigen (MTA) gene is a recently identified metastasis-associated gene which has implications in the signal transduction or regulation of gene expression. However, the expression of MTA in osteosarcoma and its potential relationship with metastasis have not been examined, forming the basis of this study.
Materials and Methods
We compared the expression levels of the MTA1 protein between 32 cases of high-grade osteosarcomas and 21 cases of low-grade osteosarcomas by immunohistochemistry. In addition, the mRNA expression levels of MTA1, 2, 3 in these osteosarcoma cell lines and control fibroblasts were evaluated by real-time quantitative PCR.
Results
MTA1 immunoreactivity was present in 81.25% of high-grade osteosarcoma specimens. Its expression was predominantly localized to the nucleus or cytoplasm of osteosarcoma cells. Thirteen (86.6%) of 15 patients who died of osteosarcomas displayed strong MTA1 expression. Both primary bone and pulmonary metastatic lesions exhibited MTA1 expression. All low-grade osteosarcomas were negative for MTA1 except for focal weak reactivity in two cases. The tested high-grade osteosarcoma cell lines showed marked amplification of MTA1 and MTA2 mRNA compared to control cells.
Conclusion
These results suggest that MTA might be involved in the progression of high-grade osteosarcoma, particularly in hematogenous metastasis of osteosarcoma.
Keywords: Metastatic tumor antigen (MTA), Osteosarcoma
INTRODUCTION
Distinguishing conventional high-grade osteosarcoma from central low-grade osteosarcoma represents an important clinical issue. Distinct sets of genes and proteins dictate the progression from precursor lesions to localized disease and finally to metastatic disease (1). Identifying and characterizing key genes that regulate the metastatic ability of cancer may help identify which tumors are on an aggressive path from the outset and treat them adequately before the development of metastases. Because distant metastasis is one of the most important factors affecting prognosis, extensive efforts have been made to predict metastasis by using tumor metastasis-associated genes or proteins (2,3). Recently, a potential metastasis-associated gene and its product, the metastatic tumor antigen 1 (MTA1), were identified; this gene has been found to be overexpressed in a variety of cancers (4~7).
MTA1 is a component of the nucleosome remodeling and histone deacetylation (NuRD) complex, which is associated with ATP-dependent chromatin remodeling and histone deacetylase activity (7). MTA1 functions in conjugation with other components of NuRD to mediate transcriptional repression and to facilitate the association of repressor molecules with chromatin (8). MTA1 is normally expressed only at low levels in various tissues (7). MTA1 enhances migration, invasion, and anchorage-independent survival of immortalized human keratinocytes (9). Elevated MTA1 levels in breast cancer appear to enhance metastasis, increase mammary cell motility, and potentiate growth; MTA1 may, therefore, be an indicator for assessing the potential aggressiveness of various tumors (7).
The expression of MTA1 in osteosarcoma and its potential relationship to pulmonary metastasis is unknown. In the present study, we examined MTA1 expression and its relationship to pulmonary metastasis in surgical specimens of human osteosarcomas. We also examined expression of the MTA1, 2, 3 mRNA in osteosarcoma cell lines.
MATERIALS AND METHODS
1) Patients and samples
Fifty-three formalin-fixed, paraffin embedded surgical specimens of osteosarcoma were obtained from the files of Kyung Hee University Hospital (Seoul, Korea) and Rizzoli Institute (Bologna, Italy). The protocol was reviewed and approved by the Institutional Review Board in Kyung Hee University Hospital. Thirty-two cases were conventional high-grade osteosarcomas and two of these included pulmonary metastatic lesions. We selected samples that were obtained prior to chemotherapy. Twenty-one cases were central low-grade osteosarcomas. Two pathologists (YKP and FB) reviewed all the cases used in this study.
2) Immunohistochemistry
The avidin-biotin complex method was performed on 4 µm thick tissue sections for immunohistochemical analysis. Sections were deparaffinized with xylene for 15 min. Sections were incubated (1:100 dilution) with the goat polyclonal antibody directed against MTA1 (C-17) (Santa Cruz Biotechnology Inc., Santa Cruz, CA) for 30 min at room temperature. Both positive and negative controls were used in each experiment. The results were expressed according to a semiquantitative scale following the criteria listed below: 0 = complete absence of tumor cells stained positive; 1+ = 1~30% of the cells stained positive; 2+ = 31~75% of the cells stained positive; 3+ = 76~100% of the cells stained positive. In each case, 10 high power fields of representative areas were counted.
3) RNA isolation from cell lines
Total RNA was extracted from three different human high-grade osteosarcoma cell lines (MG63, SJSA-1, HS3.7) and control fibroblasts from skin primary cultures using the RNeasy kit (Qiagen, Santa Clarita, CA). Cells were cultured in RPMI 1640 and DMEM media (Gibco BRL, Gaithersburg, MD) supplemented with 10% fetal bovine serum. Each sample (30 mg) was homogenized with 600 µl RLT buffer by passing through 20 gauge needles. The supernatant was transferred to a tube, an equal volume of 70% alcohol added, and the solution thoroughly mixed. This was followed by three cycles of centrifugation for 15 s. The RNA pellet was suspended in 30 µl RNase-free water and frozen at -70℃.
4) cDNA synthesis
All cDNA was synthesized using the 1st strand cDNA synthesis kit (Roche diagnostics GmbH, Mannheim, Germany). All reactions were carried out in a total volume of 20 µl per capillary. Each reaction mixture contained 2.0 µl of 10X buffer, 4.0 µl of 25 mM MgCl2, 2.0 µl of dNTP, 2.0 µl of oligo-dT primer, 1.0 µl of RNasin, 0.8 µl of AMV reverse transcriptase, 1.0 µl of RNA and sterile water. The cDNA synthesis conditions were 25℃ for 10 min, 42℃ for 60 min, and 99℃ for 5 min, using a thermal cycler (9700, Applied Biosystems, Foster City, CA).
5) Oligonucleotide primers
The sequences of the primers used in this study are shown in Table 1. Primers were designed using Primer3 software and produced with a DNA synthesizer (Polygen GmbH, Mannheim, Germany). All primers were purified by HPLC (Waters, Milford, MA).
Table 1.
Oligonucleotide primers used in this study
6) Real time PCR
PCR products were amplified and quantified by SYBR green intercalation using a LightCycler system (Roche diagnostics GmbH, Mannheim, Germany). All reactions were carried out in a total volume of 20 µl per capillary. Each reaction mixture contained: 2.0 µl of 10X buffer (50 mM KCl, 10 mM Tris-HCl (pH 8.3), and 0.001% gelatin), 2.0 µl of 2.5 mM MgCl2, 1.6 µl of 2.5 mM dNTP, 1.0 µl of primer (10 pmol/µl), 0.4 µl of Amplitaq (5 U/µl), 1.0 µl (10,000 fold dilution) of SYBR Green I (Molecular Probes, Eugene, OR, USA), 1.0 µl of DNA (100 ng/µl), and 10.0 µl of sterile water. The MTA1, 2, 3 amplification reaction conditions were as follows: 1 cycle at 95℃ for 0 s, 40 cycles at 60℃ for 5 s, and 72℃ for 12 s. The β-actin amplification reaction conditions were as follows: 1 cycle at 95℃ for 0 s, 40 cycles at 60℃ for 5 s, and 7℃ for 12 s. Melting curves were generated after the amplification procedure. Each of the tissue samples was analyzed in duplicates.
7) Data evaluation
The LightCycler apparatus measured the fluorescence of each sample at the end of the annealing step of every cycle. After proportional background adjustment, the fit point method was used to determine the cycle in which the log-linear signal was distinguishable from the background. That cycle number was used as the crossing point value. LightCycler software (version 3) produced a standard curve by measuring the crossing point of each standard and plotting them against the logarithmic values of the concentrations. The MTA1, 2, 3 and β-actin gene were cloned in the pGEM-T vector (Promega Corp., Madison, WI) for use in creating the standard curves. Ten-fold serial dilutions of known plasmid DNA concentrations were made. The gene copy number of MTA1, 2, 3 was normalized by comparison with the β-actin housekeeping gene.
RESULTS
1) Follow up data
Follow up data were available from seventeen patients of high-grade osteosarcomas. Fifteen patients died with pulmonary metastases between 1 to 50 months after diagnosis. The other two patients survived for 14 and 15 years. Among twenty-one patients of central low-grade osteosarcomas, ten cases were grade 1 and the other 11 cases were grade 2. The mean follow-up period was 80 months (range 12~158 months) in low-grade osteosarcoma patients. All the patients showed no evidence of disease at the time of follow-up.
2) Analysis of MTA protein expression in osteosarcomas
Immunohistochemistry showed that MTA1 was present in the nucleus and cytoplasm of osteosarcoma cells. Immunoreactivity was absent in benign stromal cells. Twenty-six cases (81.25%) among the 32 conventional high-grade osteosarcomas analysed showed increased staining for MTA1. The degree of overexpression was moderate (2+) in 9 cases (28.12%) and strong (3+) in 17 cases (53.12%). The immunostaining was diffuse in pattern. Among the 15 patients who died of osteosarcomas, MTA1 expression was noted in 13 of these patients (86.6%). There were no correlations between the degree of MTA1 expression and survival duration of these patients. Paraffin blocks from the pulmonary metastasis of two patients who died of it were available; all two patients exhibited MTA1 expression in both primary bone and pulmonary metastatic lesions (Fig. 1, 2). In contrast, the immunostaining for MTA1 was absent in 19 cases (90.47%) of low-grade osteosarcoma (Fig. 3). Two cases (9.52%) showed focal weak reactivity for MTA1. Histology revealed regular bony trabeculae and interlacing pattern of spindle cells, simulating either parosteal osteosarcoma or fibrous dysplasia.
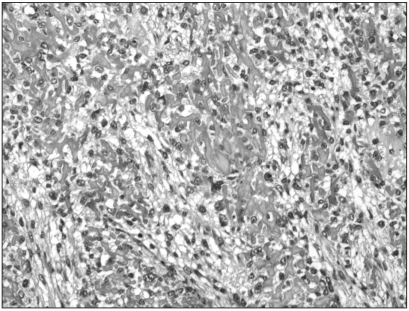
Fig. 1.
Immunohistochemistry for MTA1 in primary bone lesion of conventional high-grade osteosarcoma. Diffuse, strong expression was noted in the nucleus and cytoplasm of irregular pleomorphic tumor cells (original magnification, ×200).
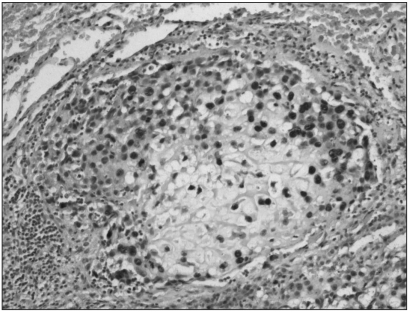
Fig. 2.
Immunohistochemistry for MTA1 in pulmonary metastatic tumor sample of high-grade osteosarcoma (original magnification, ×200).
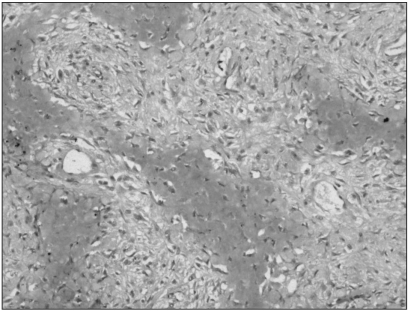
Fig. 3.
Immunohistochemistry for MTA1 in low-grade central osteosarcoma. All the tumor cells were negative (original magnification, ×200).
3) Real-time quantitative PCR
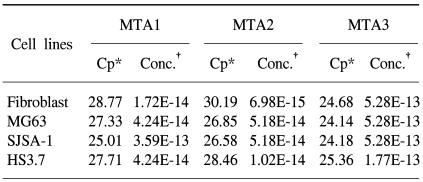
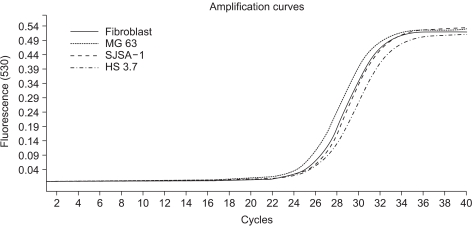
In this study, we examined three different high-grade osteosarcoma cell lines and control fibroblast for MTA1, 2, 3 mRNA. The experiment was performed five times. The crossing points and mean concentration (g/l) of each MTA, corrected using β-actin, are recorded in Table 2. All three high-grade osteosarcoma cell lines exhibited an average 8-fold and 5-fold higher amplification of MTA1 and MTA2 mRNA compared to the control, respectively. The amplification curves for the MTA1 mRNA are shown in Fig. 4. However, MTA3 mRNA level was similar in osteosarcoma cell lines and control fibroblasts.
Table 2.
Crossing points and mean concentration of each MTA mRNA in tested cell lines
*crossing points, †mean concentration (g/l).
Fig. 4.
Amplification curves for the MTA1 mRNA in osteosarcoma cell lines and control fibroblast by real-time quantitative PCR. All three osteosarcoma cell lines showed an average 8-fold higher amplification of MTA1 mRNA compared to the control.
DISCUSSION
The goal of this study was to characterize MTA expression in a wide range of osteosarcoma samples and to determine its role as a putative biomarker for osteosarcoma progression. This study is significant in that MTA was not previously characterized in osteosarcoma and that its potential as a biomarker was not determined. In this study, we have shown that MTA1 has a greater expression level in conventional high-grade osteosarcoma compared to central low-grade osteosarcoma by immunohistochemistry. MTA1 protein is strongly overexpressed in 81.25% of all examined conventional high-grade osteosarcoma. MTA1 expression was noted in most patients who died of osteosarcomas. Both primary bone and pulmonary metastatic lesions exhibited MTA1 expression. Alterations on the mRNA level, as shown through quantitative RT-PCR, correspond to this aberrant expression of MTA1. In addition, we also exhibited the amplification of MTA2 mRNA in all osteosarcoma cell lines tested. However, MTA3 mRNA levels were similar in osteosarcoma cell lines and control fibroblasts. Our data suggest that the expression of the MTA1 and MTA2 gene is associated with the progression of osteosarcoma. To clarify the role of MTA in the progression of osteosarcoma, further studies, especially those in a large scale, are required.
Our study is in agreement with previous data which show overexpression of MTA1 mRNA in human advanced cancers, including colorectal (5), breast (4), gastric (5), pancreatic (6), and liver cancers (10). Its expression was found to be associated with progression in solid cancers of various organs and cancer cell lines with high invasive potential (7) and, thus, believed to play a role in cancer progression to the metastatic state. Toh et al. (5) demonstrated that overexpression of MTA1 mRNA was correlated to the depth of invasion and lymph node metastasis in gastric and colorectal carcinomas as well as esophageal carcinomas. Similar results showing the relationship between MTA1 mRNA expression and invasion/metastasis were also reported in lung cancer and thymoma (4,11). Hofer et al. (12) found that metastatic prostate cancer displayed significantly higher mean MTA1 protein expression intensity and percentage of tissue cores staining positive for MTA1 compared to clinically localized prostate cancer or benign prostate tissue.
MTAs represent a rapidly growing novel gene family. At present, there are three different known genes (MTA1, MTA2, and MTA3) and six reported isoforms. Although some MTA1 protein was found in the cytoplasm, the vast majority of MTA1 protein was localized in the nucleus. MTA1, MTA2, and MTA3 are components of the NuRD complex, which is associated with adenosine triphosphate-dependent chromatin remodeling and transcriptional regulation (8). Toh et al. reported that the MTA1 protein is associated with histone deacetylation in vitro (13) and in clinical sample of human cancers (14). The MTA1 protein may serve multiple functions in cellular signaling, chromosome remodeling and transcription processes that are important in the progression, invasion and growth of metastatic epithelial cells (15). The overexpression of MTA1 protein and acetylation level of histone H4 protein, both of which are closely related, might be useful predictors of malignant potential of esophageal squamous cell carcinomas (14). Zhang et al. (16) reported that a protein similar to MTA1 (known as MTA2) is also a component of the NuRD complex and that MTA2 is highly expressed in rapidly dividing cells and in cervical cancer tissue, indicating a possible relationship between MTA2 expression and cellular proliferation. In addition to the MTA1/MTA2 proteins, NuRD complex contains several factors that have been reported to be associated with malignancy (17). It will be of great interest to identify the target genes of the NuRD complex and molecular analyses of the NuRD complex may provide us with important new information on carcinogenesis and cancer progression.
CONCLUSIONS
This study is the first to characterize MTA expression in conventional high-grade osteosarcomas compared to central low-grade osteosarcomas and suggests that MTA expression may be associated with the metastasis in these tumors.
References
- 1.DeMarzo AM, Nelson WG, Isaacs WB, Epstein JI. Pathological and molecular aspects of prostate cancer. Lancet. 2003;361:955–964. doi: 10.1016/S0140-6736(03)12779-1. [DOI] [PubMed] [Google Scholar]
- 2.Dear TN, Ramshaw IA, Kefford RF. Differential expression of a novel gene, WDNM1, in nonmetastatic rat mammary adenocarcinoma cells. Cancer Res. 1988;48:5203–5209. [PubMed] [Google Scholar]
- 3.Phillips SM, Bendall AJ, Ramshaw IA. Isolation of gene associated with high metastatic potential in rat mammary adenocarcinomas. J Natl Cancer Inst. 1990;82:199–203. doi: 10.1093/jnci/82.3.199. [DOI] [PubMed] [Google Scholar]
- 4.Sasaki H, Moriyama S, Nakashima Y, Kobayashi Y, Yukiue H, Kaji M, et al. Expression of the MTA1 mRNA in advanced lung cancer. Lung Cancer. 2002;35:149–154. doi: 10.1016/s0169-5002(01)00329-4. [DOI] [PubMed] [Google Scholar]
- 5.Toh Y, Oki E, Oda S, Tokunaga E, Ohna S, Maehara Y, et al. Overexpression of the MTA1 gene in gastrointestinal carcinomas: correlation with invasion and metastasis. Int J Cancer. 1997;74:459–463. doi: 10.1002/(sici)1097-0215(19970822)74:4<459::aid-ijc18>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 6.Iguchi H, Imura G, Toh Y, Ogata Y. Expression of MTA1, a metastasis-associated gene with histone deacetylase activity in pancreatic cancer. Int J Oncol. 2000;16:1211–1214. doi: 10.3892/ijo.16.6.1211. [DOI] [PubMed] [Google Scholar]
- 7.Toh Y, Pencil SD, Nicolson GL. A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. J Biol Chem. 1994;269:22958–22963. [PubMed] [Google Scholar]
- 8.Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 9.Mahoney MG, Simpson A, Jost M, Noe M, Kari C, Pepe D, et al. Metastasis-associated protein (MTA) 1 enhances migration, invasion, and anchorage-independent survival of immortalized human keratinocytes. Oncogene. 2002;21:2161–2170. doi: 10.1038/sj.onc.1205277. [DOI] [PubMed] [Google Scholar]
- 10.Moon WS, Chang K, Tarnawski AS. Overexpression of metastatic tumor antigen 1 in hepatocellular carcinoma: Relationship to vascular invasion and estrogen receptor-α. Hum Pathol. 2004;35:424–429. doi: 10.1016/j.humpath.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Sasaki H, Yukiue H, Kobayashi Y, Nakashima Y, Kaji M, Fukai I, et al. Expression of the MTA1 mRNA in thymoma patients. Cancer Lett. 2001;174:159–163. doi: 10.1016/s0304-3835(01)00704-2. [DOI] [PubMed] [Google Scholar]
- 12.Hofer MD, Kuefer R, Varambally S, Li H, Ma J, Shapiro GI, et al. The role of metastasis-associated protein 1 in prostate cancer progression. Cancer Res. 2004;64:825–829. doi: 10.1158/0008-5472.can-03-2755. [DOI] [PubMed] [Google Scholar]
- 13.Toh Y, Kuninaka S, Endo K, Oshiro T, Ikeda Y, Nakashima H, et al. Molecular analysis of a candidate metastasis-associated gene, MTA1: possible interaction with histone deacetylase 1. J Exp Clin Cancer Res. 2000;19:105–111. [PubMed] [Google Scholar]
- 14.Toh Y, Ohga T, Endo K, Adachi E, Kusumoto H, Haraguchi M, et al. Expression of the metastasis-associated MTA1 protein and its relationship to deacetylation of the histone H4 in esophageal squamous cell carcinomas. Int J Cancer. 2004;110:362–367. doi: 10.1002/ijc.20154. [DOI] [PubMed] [Google Scholar]
- 15.Nicolson G, Nawa A, Toh Y, Taniguchi S, Nishimori K, Moustafa A. Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation and nuclear regulation. Clin Exp Metastasis. 2003;20:19–24. doi: 10.1023/a:1022534217769. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Leroy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]