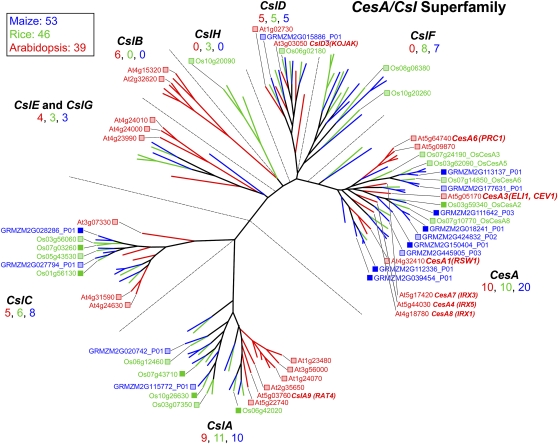
Figure 3.
Genes of the CesA/Csl superfamily. At least three CesA genes are coexpressed during primary wall formation, and mutants in each of them, AtCesA1 (rsw1; Arioli et al., 1998), AtCesA6 (procuste; Fagard et al., 2000), and AtCesA3 (cev1; Ellis et al., 2002; eli1; Caño-Delgado et al., 2003), result in cellulose deficiencies, indicating that all three are essential for cellulose synthesis. The irregular xylem mutants AtCesA8 (irx1), AtCesA7 (irx3), and AtCesA4 (irx5) are deficient in cellulose synthesis specifically in secondary walls (Taylor et al., 2003). The root-hairless mutant kojak was traced to a mutation in the CslD3 gene proposed to be a cellulose synthase in these tip-growing cells (Favery et al., 2001). Rice CslD1 (Kim et al., 2007) and maize CslD5 are apparent orthologs, as mutations in each result in the reduced root hair phenotype. Heterologous expression of the Arabidopsis CslA9 in Drosophila cells in culture confirmed the role of this gene in mannan synthesis (Liepman et al., 2005). Expression of a barley CslF gene in Arabidopsis resulted in the de novo appearance of epitopes of the mixed-linkage (1→3),(1→4)-β-d-glucan (Burton et al., 2006), and characterization of a CslC coexpressed with a xyloglucan-specific xylosyl transferase in Pichia resulted in the synthesis of extended glucan polymers (Cocuron et al., 2007). Color scheme and dendrogram labeling are as described in the legend of Figure 1. See http://cellwall.genomics.purdue.edu/families/2-1/ for accession numbers of all genes in this superfamily.