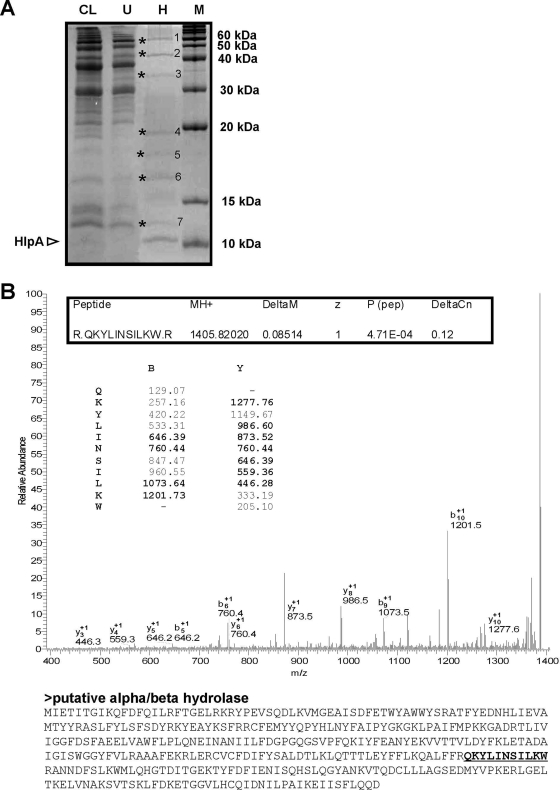
FIG. 5.
(A) SDS-PAGE analysis of a total S. gallolyticus cell lysate (CL) and the respective proteins that remain unbound (U) after incubation with immobilized heparin or have heparin affinity (H). The position of endogenous HlpA is indicated. Unknown heparin-binding proteins 1 to 7 are indicated with asterisks. M, molecular mass marker. (B) Identification of protein band 2 as a putative alpha/beta hydrolase of S. gallolyticus. Band 2, a putative heparin-binding protein (indicated in panel A), was excised from the gel and digested with trypsin to allow tandem mass spectrometry and peptide mass fingerprinting. The tandem mass spectrometry spectrum of peptide “QKYLINSILKW,” m/z 1,405.82, is shown. The theoretical series of b ions produced from cleavage of the amide bond and y ions produced by cleavage of the amide bond are indicated, with the actual identified ions printed in bold. The theoretical molecular mass of the corresponding putative alpha/beta hydrolase is 43.7 kDa, which is in-line with its mobility by SDS-PAGE (see panel A).