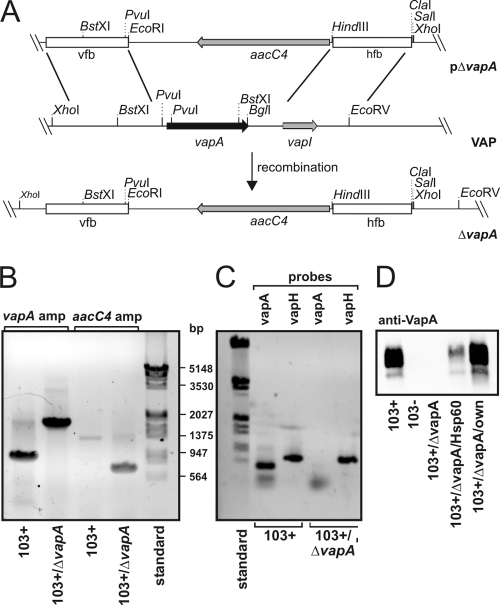
FIG. 1.
Genetic strategy to create R. equi 103+/ΔvapA. (A) Schematic representation of the creation of R. equi 103+/ΔvapA. Briefly, linearized plasmid pΔvapA was electroporated into 103+ and, after double crossover at the vfb and hfb sites, the entire vapA gene region on the virulence plasmid was replaced by the aacC4 apramycin resistance gene. “VAP” denotes the situation as is found with the relevant section of the virulence-associated plasmid, and ΔvapA describes the situation in the replacement mutant. (B) Agarose gel stained with ethidium bromide. Plasmid DNA isolated from 103+ and from 103+/ΔvapA was used as a template for PCRs with primers to up- and downstream regions of the vapA gene (vapA probes 1 and 2) yielding fragments of 900 bp in the presence of vapA and 1,800 bp in its absence (corresponding to the aacC4 gene with flanking regions of vapA). Primer pair aacC4-F/aacC4-R was used to amplify a fragment of 743 bp in the open reading frame of the apramycin resistance gene with the nucleotide sequence analyzed from pVKT173. A molecular weight marker standard (bp) is shown on the right. (C) The virulence plasmids from 103+ or 103+/ΔvapA were used in PCRs with primers that amplify either the vapA gene as described above or the vapH gene (oligonucleotides vapH-F/-R), which is also located on the VAP. Although vapA is present only in 103+, vapH is present in the wild type and mutant, demonstrating the presence of the VAP in either strain. (D) Immunoblot of R. equi proteins developed with a monoclonal antibody to VapA. Equivalent cell numbers of R. equi overnight cultures were used.