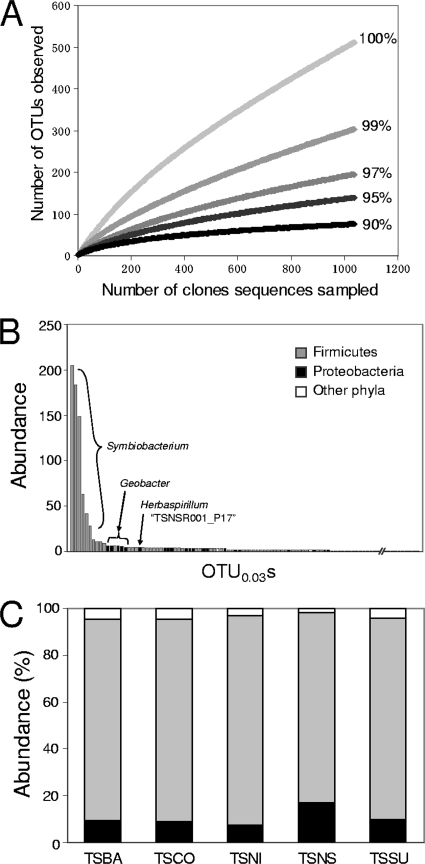
FIG. 2.
Diversity in the clone libraries. (A) Rarefaction curve analysis performed with sequenced clones obtained from the unincubated control (TSBA sample). Five different sequence dissimilarity values (0, 1, 3, 5, and 10%) were used to define OTUs. Similar results were observed with clones obtained from the other samples (data not shown). (B) Rank distribution analysis performed with OTU0.03s obtained from the TSBA sample. The most abundant OTU0.03s belonged to the Symbiobacterium clade (Firmicutes). (C) Distribution of various phyla in the clone libraries. The other phyla include Acidobacteria, Actinobacteria, Bacteroidetes, the BRC1 clade, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Nitrospira, the OD1 clade, Planctomycetes, Verrucomicrobia, and unclassified bacteria.